5GUY
 
 | | Structural and functional study of chuY from E. coli CFT073 strain | | Descriptor: | Uncharacterized protein chuY | | Authors: | Kim, H, Choi, J, HA, S.C, Chaurasia, A.K, Kim, D, Kim, K.K. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural and functional study of ChuY from Escherichia coli strain CFT073
Biochem. Biophys. Res. Commun., 482, 2017
|
|
1L1J
 
 | | Crystal structure of the protease domain of an ATP-independent heat shock protease HtrA | | Descriptor: | heat shock protease HtrA | | Authors: | Kim, D.Y, Kim, D.R, Ha, S.C, Lokanath, N.K, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2002-02-18 | | Release date: | 2003-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Protease Domain of a Heat-shock Protein HtrA from Thermotoga maritima
J.BIOL.CHEM., 278, 2003
|
|
5WUQ
 
 | | Crystal structure of SigW in complex with its anti-sigma RsiW, a zinc binding form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW, ZINC ION | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
5WUR
 
 | | Crystal structure of SigW in complex with its anti-sigma RsiW, an oxdized form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
1PCV
 
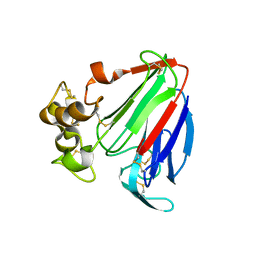 | | Crystal structure of osmotin, a plant antifungal protein | | Descriptor: | osmotin | | Authors: | Min, K, Ha, S.C, Yun, D.-J, Bressan, R.A, Kim, K.K. | | Deposit date: | 2003-05-16 | | Release date: | 2004-02-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of osmotin, a plant antifungal protein
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3WT4
 
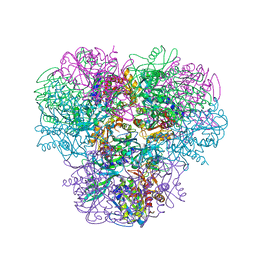 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | CARBONATE ION, Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-04-07 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
3ZPJ
 
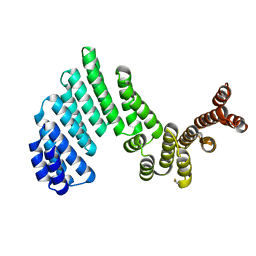 | | Crystal structure of Ton1535 from Thermococcus onnurineus NA1 | | Descriptor: | TON_1535 | | Authors: | Jeong, J.H, kim, Y.G. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structure of the Hypothetical Protein Ton1535 from Thermococcus Onnurineus Na1 Reveals Unique Structural Properties by a Left-Handed Helical Turn in Normal Alpha-Solenoid Protein.
Proteins, 82, 2014
|
|
4UY5
 
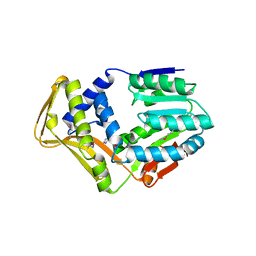 | |
4UY7
 
 | |
4UY6
 
 | |
5HZT
 
 | | Crystal structure of Dronpa-Cu2+ | | Descriptor: | COPPER (II) ION, Fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HZU
 
 | | Crystal structure of Dronpa-Ni2+ | | Descriptor: | Fluorescent protein Dronpa, NICKEL (II) ION | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HZS
 
 | | Crystal structure of Dronpa-Co2+ | | Descriptor: | COBALT (II) ION, Fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5H6Z
 
 | | Crystal structure of Set7, a novel histone methyltransferase in Schizossacharomyces pombe | | Descriptor: | AMMONIUM ION, DI(HYDROXYETHYL)ETHER, SET domain-containing protein 7, ... | | Authors: | Mevius, D.E.H.F, Shen, Y, Morishita, M, Carrozzini, B, Caliandro, R, di Luccio, E. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Set7 Is a H3K37 Methyltransferase in Schizosaccharomyces pombe and Is Required for Proper Gametogenesis.
Structure, 27, 2019
|
|
7C41
 
 | | KRAS G12V and H-REV107 peptide complex | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, HRAS-like suppressor 3, ... | | Authors: | Han, C.W, Jeong, M.S, Jang, S.B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | A H-REV107 Peptide Inhibits Tumor Growth and Interacts Directly with Oncogenic KRAS Mutants.
Cancers (Basel), 12, 2020
|
|
7C3Z
 
 | |
7C40
 
 | | MgGDP bound KRAS G12V | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Han, C.W, Jeong, M.S, Jang, S.B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.516 Å) | | Cite: | A H-REV107 Peptide Inhibits Tumor Growth and Interacts Directly with Oncogenic KRAS Mutants.
Cancers (Basel), 12, 2020
|
|
6JHK
 
 | |
4KMF
 
 | |
3JUJ
 
 | |
3JUK
 
 | |
5Y6B
 
 | |
5GPG
 
 | | Co-crystal structure of the FK506 binding domain of human FKBP25, Rapamycin and the FRB domain of human mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP3, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Lee, H.B, Lee, S.Y, Rhee, H.W, Lee, C.W. | | Deposit date: | 2016-08-02 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proximity-Directed Labeling Reveals a New Rapamycin-Induced Heterodimer of FKBP25 and FRB in Live Cells
Acs Cent.Sci., 2, 2016
|
|
5ZU1
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*AP*GP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*TP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-05 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5ZUO
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
