5NN7
 
 | | KSHV uracil-DNA glycosylase, apo form | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | Deposit date: | 2017-04-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NNU
 
 | | KSHV uracil-DNA glycosylase, product complex with dsDNA exhibiting duplex nucleotide flipping | | Descriptor: | DNA, DNA containing an abasic site, Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Barrett, T, Savva, R. | | Deposit date: | 2017-04-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NNH
 
 | | KSHV uracil-DNA glycosylase, apo form | | Descriptor: | SULFATE ION, Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | Deposit date: | 2017-04-09 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
8AIL
 
 | | Bacillus phage VMY22 p56 in complex with Bacillus weidmannii Ung | | Descriptor: | Bacillus phage VMY22 p56, GLYCEROL, IODIDE ION, ... | | Authors: | Muselmani, W, Bagneris, C, Savva, R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
2W2K
 
 | | Crystal structure of the apo forms of Rhodotorula graminis D- mandelate dehydrogenase at 1.8A. | | Descriptor: | D-MANDELATE DEHYDROGENASE | | Authors: | Vachieri, S.G, Cole, A.R, Bagneris, C, Baker, D.P, Fewson, C.A, Basak, A.K. | | Deposit date: | 2008-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Apo and Holo Forms of Rhodotorula Graminis D(-)-Mandelate Dehydrogenase
To be Published
|
|
7PDJ
 
 | | R12E vFLIP mutant | | Descriptor: | FLICE inhibitory protein | | Authors: | Barrett, T.E. | | Deposit date: | 2021-08-05 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Mechanistic insights into the activation of the IKK kinase complex by the Kaposi's sarcoma herpes virus oncoprotein vFLIP.
J.Biol.Chem., 298, 2022
|
|
1XSF
 
 | | Solution structure of a resuscitation promoting factor domain from Mycobacterium tuberculosis | | Descriptor: | Probable resuscitation-promoting factor rpfB | | Authors: | Cohen-Gonsaud, M, Barthe, P, Henderson, B, Ward, J, Roumestand, C, Keep, N.H. | | Deposit date: | 2004-10-19 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes
Nat.Struct.Mol.Biol., 12, 2005
|
|
5LDE
 
 | | Crystal structure of a vFLIP-IKKgamma stapled peptide dimer | | Descriptor: | Immunoglobulin G-binding protein G,Viral FLICE protein, Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, ... | | Authors: | Barrett, T. | | Deposit date: | 2016-06-24 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | IKK gamma-Mimetic Peptides Block the Resistance to Apoptosis Associated with Kaposi's Sarcoma-Associated Herpesvirus Infection.
J. Virol., 91, 2017
|
|
3V4R
 
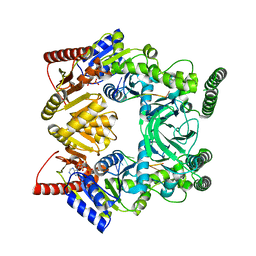 | | Crystal structure of a UvrB dimer-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA: 5 -TACTGTTT-3, UvrABC system protein B | | Authors: | Webster, M.P.J, Jukes, R, Barrett, T. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the UvrB dimer: insights into the nature and functioning of the UvrAB damage engagement and UvrB-DNA complexes.
Nucleic Acids Res., 40, 2012
|
|
5HSW
 
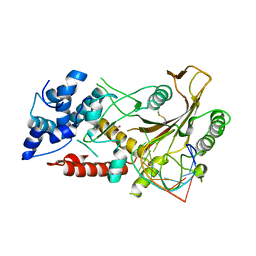 | | KSHV SOX RNA complex | | Descriptor: | ACETATE ION, ORF 37, RNA (5'-R(P*UP*CP*UP*UP*GP*AP*AP*GP*CP*AP*GP*CP*UP*UP*CP*CP*AP*G)-3') | | Authors: | Barrett, T.E. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | KSHV SOX mediated host shutoff: the molecular mechanism underlying mRNA transcript processing.
Nucleic Acids Res., 45, 2017
|
|
8AIN
 
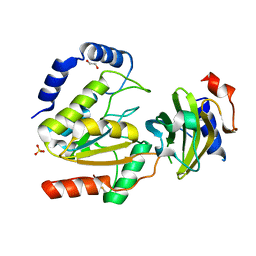 | | MCUGI SAUNG complex | | Descriptor: | 1,2-ETHANEDIOL, MCUGI, SULFATE ION, ... | | Authors: | Muselmani, W, Savva, R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
8AIM
 
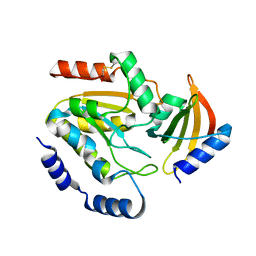 | | Ugi-2 SAUNG complex | | Descriptor: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Muselmani, W, Savva, R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
4OW1
 
 | | Crystal Structure of Resuscitation Promoting Factor C | | Descriptor: | 1,2-ETHANEDIOL, Resuscitation-promoting factor RpfC | | Authors: | Chauviac, F.X, Quay, D.H.X, Cohen-Gonsaud, M, Keep, N.H. | | Deposit date: | 2014-01-29 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The RpfC (Rv1884) atomic structure shows high structural conservation within the resuscitation-promoting factor catalytic domain.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5HVD
 
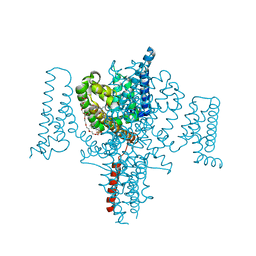 | | Full length Open-form Sodium Channel NavMs I218C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Booker, J.E, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2016-01-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The complete structure of an activated open sodium channel.
Nat Commun, 8, 2017
|
|
5HVX
 
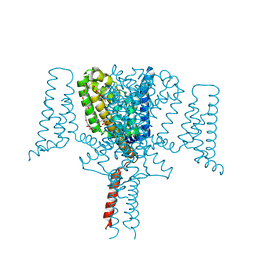 | | Full length Wild-Type Open-form Sodium Channel NavMs | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECAETHYLENE GLYCOL, ... | | Authors: | Sula, A, Booker, J.E, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2016-01-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The complete structure of an activated open sodium channel.
Nat Commun, 8, 2017
|
|
