8JYB
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 | | Descriptor: | Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
4LGI
 
 | | N-terminal truncated NleC structure | | Descriptor: | Uncharacterized protein | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LGJ
 
 | | Crystal structure and mechanism of a type III secretion protease | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6W9H
 
 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | 4-{(3R)-3-[4-(benzyloxy)phenyl]-3-[(4-fluorophenyl)sulfonyl]pyrrolidine-1-carbonyl}-1lambda~6~-thiane-1,1-dione, Nuclear receptor ROR-gamma, Steroid receptor coactivator 1 fusion | | Authors: | Sack, J.S. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6W9I
 
 | |
4OA7
 
 | | Crystal structure of Tankyrase1 in complex with IWR1 | | Descriptor: | 4-[(3aR,4R,7S,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Zhang, X, He, H. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
8EQN
 
 | |
3TWO
 
 | | The crystal structure of CAD from Helicobacter pylori complexed with NADP(H) | | Descriptor: | Mannitol dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Seo, K.H, Zhuang, N.N, Cong, C, Lee, K.H. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Unusual NADPH conformation in the crystal structure of a cinnamyl alcohol dehydrogenase from Helicobacter pylori in complex with NADP(H) and substrate docking analysis
Febs Lett., 586, 2012
|
|
5ZXD
 
 | | Crystal structure of ATP-bound human ABCF1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family F member 1 | | Authors: | Qu, L, Jiang, Y, Liu, Z.J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-07-25 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Crystal Structure of ATP-Bound Human ABCF1 Demonstrates a Unique Conformation of ABC Proteins
Structure, 26, 2018
|
|
5B6E
 
 | |
2M7T
 
 | | Solution NMR Structure of Engineered Cystine Knot Protein 2.5D | | Descriptor: | Cystine Knot Protein 2.5D | | Authors: | Cochran, F.V, Das, R. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
7KMK
 
 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, two RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KLG
 
 | | SARS-CoV-2 RBD in complex with Fab 15033 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033 heavy chain, Fab 15033 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KML
 
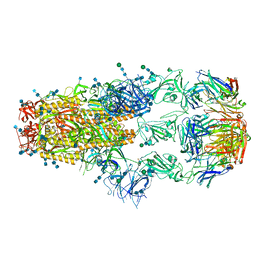 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, three RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KLH
 
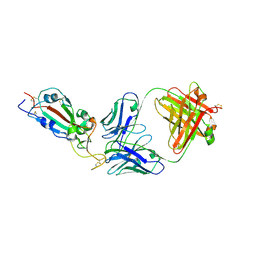 | | SARS-CoV-2 RBD in complex with Fab 15033-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, Fab 15033-7 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
5B6D
 
 | |
3C0F
 
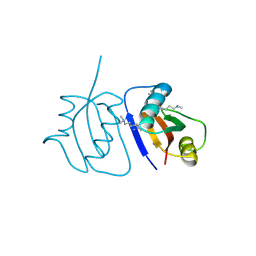 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
5J87
 
 | |
4AE0
 
 | |
3GCW
 
 | | PCSK9:EGFA(H306Y) | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Kwon, H.J. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antagonism of Secreted PCSK9 Increases Low Density Lipoprotein Receptor Expression in HepG2 Cells.
J.Biol.Chem., 284, 2009
|
|
3TS0
 
 | | Mouse Lin28A in complex with let-7f-1 microRNA pre-element | | Descriptor: | Protein lin-28 homolog A, RNA (5'-R(*GP*GP*GP*GP*UP*AP*GP*UP*GP*AP*UP*UP*UP*UP*AP*CP*CP*CP*UP*GP*GP*AP*G)-3'), ZINC ION | | Authors: | Nam, Y, Sliz, P. | | Deposit date: | 2011-09-11 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.763 Å) | | Cite: | Molecular Basis for Interaction of let-7 MicroRNAs with Lin28.
Cell(Cambridge,Mass.), 147, 2011
|
|
6MEV
 
 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
3VB7
 
 | | Crystal structure of SARS-CoV 3C-like protease with M4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, GLYCEROL, ... | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
8SUX
 
 | | Structure of E. coli PtuA hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3VB6
 
 | | Crystal structure of SARS-CoV 3C-like protease with C6Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, C6Z inhibitor | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
