6JVG
 
 | | Crystal structure of human MTH1 in complex with compound MI0639 | | Descriptor: | 5-ethyl-4-methyl-6-(morpholin-4-yl)pyrimidin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVQ
 
 | | Crystal structure of human MTH1 in complex with compound MI1025 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-morpholin-4-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JGB
 
 | | Crystal structure of barley exohydrolaseI W286F mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGQ
 
 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 2-thio-beta-sophoroside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGN
 
 | | Crystal structure of barley exohydrolaseI W434H in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
2ZE1
 
 | | X-ray structure of Bace-1 in complex with compound 6g | | Descriptor: | 3-bromo-N-[4-[1-(2-carbamimidamido-2-oxo-ethyl)-5-phenyl-pyrrol-2-yl]phenyl]benzamide, Beta-secretase 1 | | Authors: | Chopra, R, Olland, A. | | Deposit date: | 2007-12-05 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6KUF
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with glucose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-09-02 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6L1J
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with 4'-nitrophenyl thiolaminaritrioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6I28
 
 | | Crystal Structure of Cydia Pomonella PTP-2 phosphatase | | Descriptor: | CALCIUM ION, GLYCEROL, ORF98 PTP-2 | | Authors: | Huang, G, Keown, J.P, Oliver, M.R, Metcalf, P. | | Deposit date: | 2018-10-31 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of protein tyrosine phosphatase-2 from Cydia pomonella granulovirus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6JVJ
 
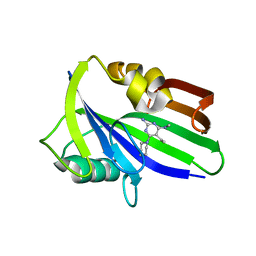 | | Crystal structure of human MTH1 in complex with compound MI1006 | | Descriptor: | 5-ethyl-N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVR
 
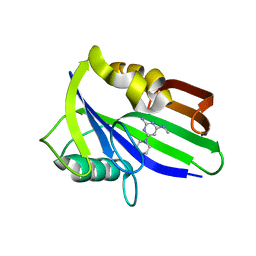 | | Crystal structure of human MTH1 in complex with compound MI1026 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
1ZCE
 
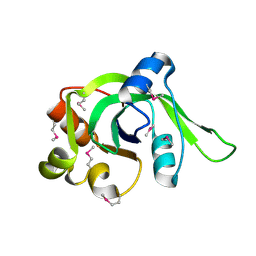 | | X-Ray Crystal Structure of Protein Atu2648 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR33. | | Descriptor: | hypothetical protein Atu2648 | | Authors: | Forouhar, F, Chen, Y, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-11 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
2ZDZ
 
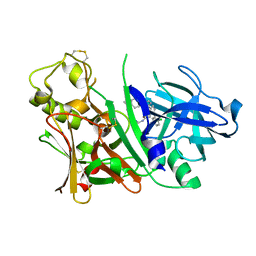 | | X-ray structure of Bace-1 in complex with compound 3.b.10 | | Descriptor: | Beta-secretase 1, N-carbamimidoyl-2-[2-(2-chlorophenyl)-5-[4-(4-ethanoylphenoxy)phenyl]pyrrol-1-yl]ethanamide | | Authors: | Chopra, R, Olland, A. | | Deposit date: | 2007-12-04 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6JO2
 
 | | Ferredoxin I from Thermosynechococcus elongatus | | Descriptor: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, ... | | Authors: | Motomura, T. | | Deposit date: | 2019-03-19 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An alternative plant-like cyanobacterial ferredoxin with unprecedented structural and functional properties.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6JGE
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with methyl 2-thio-beta-sophoroside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGC
 
 | | Crystal structure of barley exohydrolaseI W286Y mutant in complex with glucose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JG7
 
 | | Crystal structure of barley exohydrolaseI W286F in complex with methyl 2-thio-beta-sophoroside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGS
 
 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6LC5
 
 | | Crystal structure of barley exohydrolaseI W434F in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-oxane-2,3,5-triol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-11-17 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
3B6A
 
 | | Crystal structure of the Streptomyces coelicolor TetR family protein ActR in complex with actinorhodin | | Descriptor: | 2,2'-[(1R,1'R,3S,3'S)-6,6',9,9'-tetrahydroxy-1,1'-dimethyl-5,5',10,10'-tetraoxo-3,3',4,4',5,5',10,10'-octahydro-1H,1'H-8,8'-bibenzo[g]isochromene-3,3'-diyl]diacetic acid, ActR protein | | Authors: | Willems, A.R, Junop, M.S. | | Deposit date: | 2007-10-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA.
J.Mol.Biol., 376, 2008
|
|
3BJE
 
 | |
3BAX
 
 | |
3BBX
 
 | |
3BAW
 
 | | Human pancreatic alpha-amylase complexed with azide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AZIDE ION, CALCIUM ION, ... | | Authors: | Maurus, R, Brayer, G.D. | | Deposit date: | 2007-11-08 | | Release date: | 2008-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
3B6C
 
 | |
