6C4Q
 
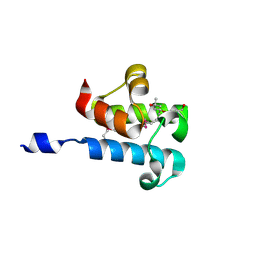 | | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Polyketide synthase Pks13 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1.16 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1-100) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To Be Published
|
|
4JB7
 
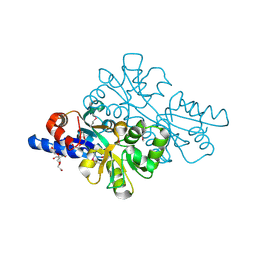 | | 1.42 Angstrom resolution crystal structure of accessory colonization factor AcfC (acfC) in complex with D-aspartic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Accessory colonization factor AcfC, D-MALATE, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Dubrovska, I, Winsor, J, Minasov, G, Shuvalova, L, Filippova, E.V, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42 Angstrom resolution crystal structure of accessory colonization factor AcfC (acfC) in complex with D-aspartic acid
To be Published
|
|
6CMZ
 
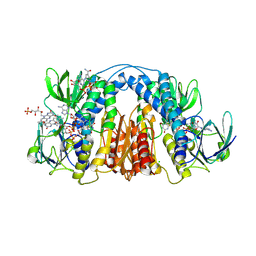 | | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D-MALATE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD.
To Be Published
|
|
4H1H
 
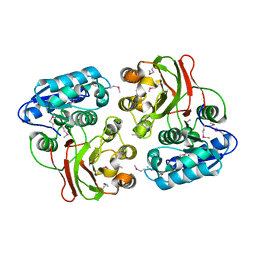 | |
4HCI
 
 | | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 from Bacillus anthracis | | Descriptor: | Cupredoxin 1, GLYCEROL | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 from Bacillus anthracis
To be Published
|
|
4HCG
 
 | | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Zinc bound from Bacillus anthracis | | Descriptor: | Cupredoxin 1, ZINC ION | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-29 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Zinc bound from Bacillus anthracis
To be Published
|
|
6BXG
 
 | | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide. | | Descriptor: | CHLORIDE ION, IODIDE ION, LEU-ILE-ALA, ... | | Authors: | Minasov, G, Shuvalova, L, Filippova, E.V, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide.
To Be Published
|
|
4K15
 
 | | 2.75 Angstrom Crystal Structure of Hypothetical Protein lmo2686 from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, Lmo2686 protein | | Authors: | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-04 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Crystal Structure of Hypothetical Protein lmo2686 from Listeria monocytogenes EGD-e
TO BE PUBLISHED
|
|
6C43
 
 | | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium. | | Descriptor: | Gamma-aminobutyraldehyde dehydrogenase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Tekleab, H, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-11 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium.
To Be Published
|
|
4IQE
 
 | | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor | | Descriptor: | Carboxyvinyl-carboxyphosphonate phosphorylmutase | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
4IQF
 
 | | Crystal Structure of Methyionyl-tRNA Formyltransferase from Bacillus anthracis | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, GLYCEROL, Methionyl-tRNA formyltransferase, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Crystal Structure of Methyionyl-tRNA Formyltransferase from Bacillus anthracis
To be Published
|
|
4IR0
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
4IR8
 
 | | 1.85 Angstrom Crystal Structure of Putative Sedoheptulose-1,7 bisphosphatase from Toxoplasma gondii | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Sedoheptulose-1,7 bisphosphatase, ... | | Authors: | Minasov, G, Ruan, J, Wawrzak, Z, Halavaty, A, Shuvalova, L, Harb, O.S, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of Putative Sedoheptulose-1,7 bisphosphatase from Toxoplasma gondii.
TO BE PUBLISHED
|
|
3IG4
 
 | | Structure of a putative aminopeptidase P from Bacillus anthracis | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Xaa-pro aminopeptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a putative aminopeptidase P from Bacillus anthracis
To be Published
|
|
5V36
 
 | | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-06 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD.
To Be Published
|
|
4FO1
 
 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, apo | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lincosamide resistance protein | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, apo
TO BE PUBLISHED
|
|
6BZ0
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD. | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.
To Be Published
|
|
6E4B
 
 | |
5VDN
 
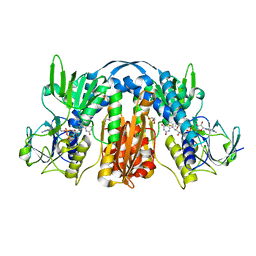 | | 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Glutathione oxidoreductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-03 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD.
To Be Published
|
|
4ERR
 
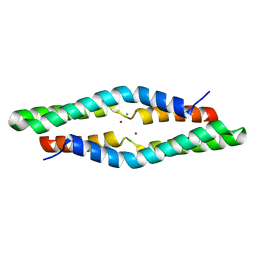 | | 1.55 Angstrom Crystal Structure of the Four Helical Bundle Membrane Localization Domain (4HBM) of the Vibrio vulnificus MARTX Effector Domain DUF5 | | Descriptor: | Autotransporter adhesin, BROMIDE ION, CHLORIDE ION | | Authors: | Minasov, G, Wawrzak, Z, Geissler, B, Shuvalova, L, Dubrovska, I, Winsor, J, Satchell, K.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Crystal Structure of the Four Helical Bundle Membrane Localization Domain (4HBM) of the Vibrio vulnificus MARTX Effector Domain DUF5.
TO BE PUBLISHED
|
|
4FBD
 
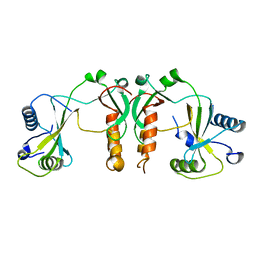 | | 2.35 Angstrom Crystal Structure of Conserved Hypothetical Protein from Toxoplasma gondii ME49. | | Descriptor: | Putative uncharacterized protein | | Authors: | Minasov, G, Ruan, J, Wawrzak, Z, Shuvalova, L, Ngo, H, Knoll, L, Milligan-Myhre, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom Crystal Structure of Conserved Hypothetical Protein from Toxoplasma gondii ME49.
TO BE PUBLISHED
|
|
3H83
 
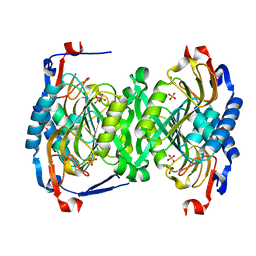 | | 2.06 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | Hypoxanthine phosphoribosyltransferase, PHOSPHATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Dubrovska, I, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 2.06 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
6DIN
 
 | | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase | | Descriptor: | SULFATE ION, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Chang, C, Jedrzejczak, R, Chhor, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-23 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase
To Be Published
|
|
4EQB
 
 | | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein PotD from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES.
TO BE PUBLISHED
|
|
5VH6
 
 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
