1QH0
 
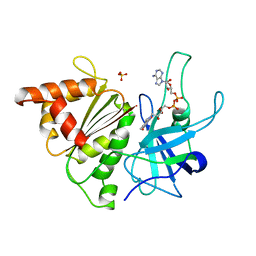 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 76 MUTATED BY ASP AND LEU 78 MUTATED BY ASP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
1QOF
 
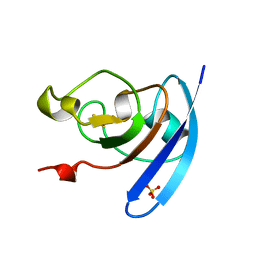 | | FERREDOXIN MUTATION Q70K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1QOG
 
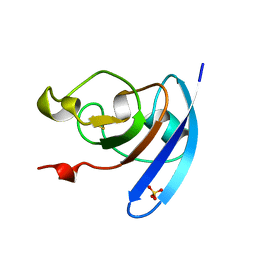 | | FERREDOXIN MUTATION S47A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
3HKK
 
 | |
1IJ8
 
 | | CRYSTAL STRUCTURE OF LITE AVIDIN-BNI COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, AVIDIN | | Authors: | Livnah, O, Huberman, T. | | Deposit date: | 2001-04-25 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chicken avidin exhibits pseudo-catalytic properties. Biochemical, structural, and electrostatic consequences.
J.Biol.Chem., 276, 2001
|
|
1IGU
 
 | |
3RGE
 
 | |
3RG3
 
 | |
1I9H
 
 | | CORE STREPTAVIDIN-BNA COMPLEX | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, STREPTAVIDIN | | Authors: | Livnah, O, Huberman, T, Wilchek, M, Bayer, E.A, Eisenberg-Domovich, Y. | | Deposit date: | 2001-03-20 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chicken avidin exhibits pseudo-catalytic properties. Biochemical, structural, and electrostatic consequences.
J.Biol.Chem., 276, 2001
|
|
3RG4
 
 | |
1IGQ
 
 | |
1AZJ
 
 | |
1AZK
 
 | |
1AZ6
 
 | |
2CA3
 
 | | Sulfite dehydrogenase from Starkeya Novella r55m mutant | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, HEME C, SULFATE ION, ... | | Authors: | Bailey, S, Kappler, U, Feng, C, Honeychurch, M.J, Bernhardt, P.V, Tollin, G, Enemark, J.H. | | Deposit date: | 2005-12-16 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for enzymatic sulfite oxidation: how three conserved active site residues shape enzyme activity.
J.Biol.Chem., 284, 2009
|
|
1AZH
 
 | |
1BJK
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH ARG 264 REPLACED BY GLU (R264E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Arg100 and Arg264 from Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal NADP+ binding and electron transfer.
Biochemistry, 37, 1998
|
|
1CJV
 
 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH BETA-L-2',3'-DIDEOXYATP, MG, AND ZN | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1CJK
 
 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH ADENOSINE 5'-(ALPHA THIO)-TRIPHOSPHATE (RP), MG, AND MN | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADENOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-16 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1CJU
 
 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH BETA-L-2',3'-DIDEOXYATP AND MG | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADENYLATE CYCLASE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-16 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1CJT
 
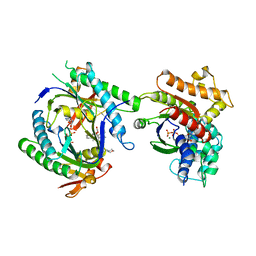 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH BETA-L-2',3'-DIDEOXYATP, MN, AND MG | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-16 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
2LJ9
 
 | |
1A4U
 
 | |
