1JZ7
 
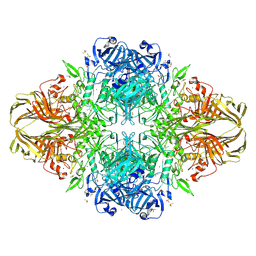 | |
1JZ3
 
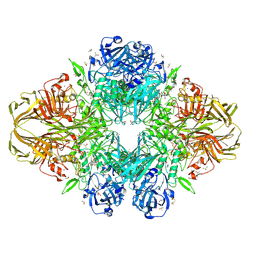 | |
1JZ5
 
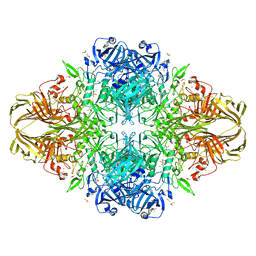 | |
1JZ6
 
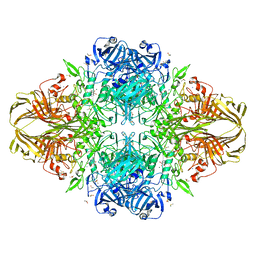 | | E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTO-TETRAZOLE | | Descriptor: | (5R, 6S, 7S, ... | | Authors: | Juers, D.H, Heightman, T.D, Vasella, A, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
1JYN
 
 | |
1JZ8
 
 | |
3PBG
 
 | | 6-PHOSPHO-BETA-GALACTOSIDASE FORM-C | | Descriptor: | 6-PHOSPHO-BETA-D-GALACTOSIDASE, SULFATE ION | | Authors: | Wiesmann, C, Schulz, G.E. | | Deposit date: | 1997-02-21 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures and mechanism of 6-phospho-beta-galactosidase from Lactococcus lactis.
J.Mol.Biol., 269, 1997
|
|
1PBG
 
 | |
2NLR
 
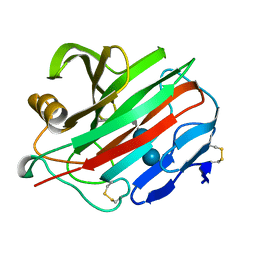 | |
2C83
 
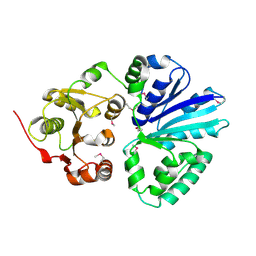 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 | | Descriptor: | HYPOTHETICAL PROTEIN PM0188 | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
2C84
 
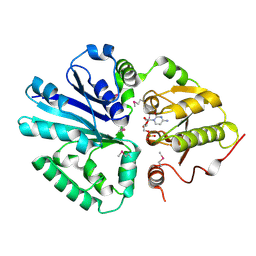 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 WITH CMP | | Descriptor: | ALPHA-2,3/2,6-SIALYLTRANSFERASE/SIALIDASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
2CXG
 
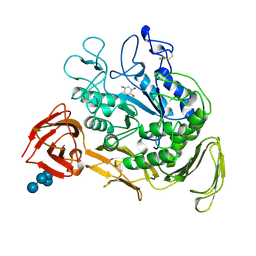 | | CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED TO THE INHIBITOR ACARBOSE | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Uitdehaag, J.C.M, Ruiterkamp, R, Dijkstra, B.W. | | Deposit date: | 1998-05-08 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases.
Biochemistry, 34, 1995
|
|
1D3C
 
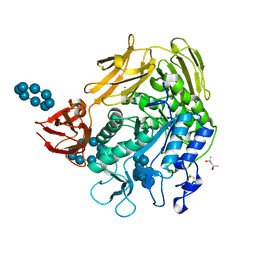 | | MICHAELIS COMPLEX OF BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE WITH GAMMA-CYCLODEXTRIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, van der Veen, B.A, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 1999-09-29 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The cyclization mechanism of cyclodextrin glycosyltransferase (CGTase) as revealed by a gamma-cyclodextrin-CGTase complex at 1.8-A resolution.
J.Biol.Chem., 274, 1999
|
|
1EO7
 
 | | BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE IN COMPLEX WITH MALTOHEXAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 2000-03-22 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structures of maltohexaose and maltoheptaose bound at the donor sites of cyclodextrin glycosyltransferase give insight into the mechanisms of transglycosylation activity and cyclodextrin size specificity.
Biochemistry, 39, 2000
|
|
1EO5
 
 | |
1CX1
 
 | | SECOND N-TERMINAL CELLULOSE-BINDING DOMAIN FROM CELLULOMONAS FIMI BETA-1,4-GLUCANASE C, NMR, 22 STRUCTURES | | Descriptor: | ENDOGLUCANASE C | | Authors: | Brun, E, Johnson, P.E, Creagh, L.A, Haynes, C.A, Tomme, P, Webster, P, Kilburn, D.G, McIntosh, L.P. | | Deposit date: | 1999-08-27 | | Release date: | 2000-04-02 | | Last modified: | 2017-02-01 | | Method: | SOLUTION NMR | | Cite: | Structure and binding specificity of the second N-terminal cellulose-binding domain from Cellulomonas fimi endoglucanase C.
Biochemistry, 39, 2000
|
|
1AXR
 
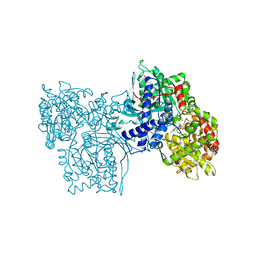 | | COOPERATIVITY BETWEEN HYDROGEN-BONDING AND CHARGE-DIPOLE INTERACTIONS IN THE INHIBITION OF BETA-GLYCOSIDASES BY AZOLOPYRIDINES: EVIDENCE FROM A STUDY WITH GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 4,5,6-TRIHYDROXY-7-HYDROXYMETHYL-4,5,6,7-TETRAHYDRO-1H-[1,2,3]TRIAZOLO[1,5-A]PYRIDIN-8-YLIUM, GLYCOGEN PHOSPHORYLASE, PHOSPHATE ION, ... | | Authors: | Oikonomakos, N.G, Heightman, T.D. | | Deposit date: | 1997-10-20 | | Release date: | 1998-01-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cooperative interactions of the catalytic nucleophile and the catalytic acid in the inhibition of beta-glycosidases. Calculations and their validation by comparative kinetic and structural studies of the inhibition of glycogen phosphorylase b.
HELV.CHIM.ACTA, 81, 1998
|
|
2PBG
 
 | | 6-PHOSPHO-BETA-D-GALACTOSIDASE FORM-B | | Descriptor: | 6-PHOSPHO-BETA-D-GALACTOSIDASE, SULFATE ION | | Authors: | Wiesmann, C, Schulz, G.E. | | Deposit date: | 1997-02-21 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures and mechanism of 6-phospho-beta-galactosidase from Lactococcus lactis.
J.Mol.Biol., 269, 1997
|
|
1GPA
 
 | |
1GPB
 
 | |
