1QAK
 
 | | THE ACTIVE SITE BASE CONTROLS COFACTOR REACTIVITY IN ESCHERICHIA COLI AMINE OXIDASE : X-RAY CRYSTALLOGRAPHIC STUDIES WITH MUTATIONAL VARIANTS | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E, McPherson, M.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: x-ray crystallographic studies with mutational variants.
Biochemistry, 38, 1999
|
|
1R0F
 
 | | Gallium-substituted rubredoxin | | Descriptor: | GALLIUM (III) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R0H
 
 | | cobalt-substituted rubredoxin | | Descriptor: | COBALT (II) ION, Rubredoxin | | Authors: | Maher, M, Cross, M, Wilce, M.C.J, Guss, J.M, Wedd, A.G. | | Deposit date: | 2003-09-22 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R0N
 
 | | Crystal Structure of Heterodimeric Ecdsyone receptor DNA binding complex | | Descriptor: | Ecdsyone Response Element, Ecdysone Response Element, Ecdysone receptor, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the heterodimeric Ecdysone Receptor DNA-binding complex
Embo J., 22, 2003
|
|
1QKT
 
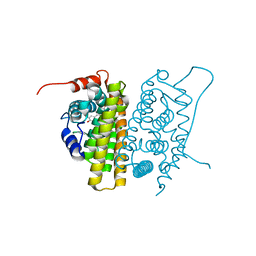 | | MUTANT ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Mutant Heralpha Ligand- Binding Domain Reveals Key Structural Features for the Mechanism of Partial Agonism
J.Biol.Chem., 276, 2001
|
|
2AP2
 
 | |
7OI2
 
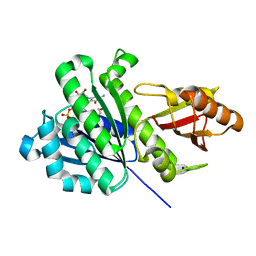 | | SaFtsz complexed with GDP (NaCl purification) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu Morales, J.M. | | Deposit date: | 2021-05-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7SG3
 
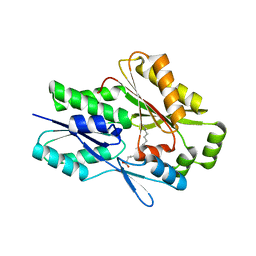 | | The X-ray crystal structure of the Staphylococcus aureus Fatty Acid Kinase B1 mutant A121I-A158L to 2.35 Angstrom resolution (Open form chain A, Palmitate bound) | | Descriptor: | Fatty Acid Kinase B1, PALMITIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Rock, C.O, White, S.W. | | Deposit date: | 2021-10-04 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7S4Z
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Proteinase K | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S4W
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S4Y
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7SCL
 
 | | The X-ray crystal structure of Staphylococcus aureus Fatty Acid Kinase B1 (FakB1) mutant R173A in complex with Palmitate to 1.60 Angstrom resolution | | Descriptor: | Fatty Acid Kinase B1, GLYCEROL, PALMITIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7S50
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Phycocyanin | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, CHLORIDE ION, ... | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S4R
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - alpha Spectrin-SH3 domain | | Descriptor: | Spectrin alpha chain, non-erythrocytic 1 | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7S56
 
 | | Sortase A from Streptococcus agalactiae, residues 79-247 | | Descriptor: | Class A sortase | | Authors: | Gao, M, Kodama, H.M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7S54
 
 | | Sortase A from Streptococcus agalactiae with the deltaN188 b7-b8 loop sequence from Staphylococcus aureus Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Gao, M, Kodama, H.M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7S57
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence of Enterococcus faecalis Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Svendsen, J.E, Johnson, D.A, Gao, M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7S53
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence from Listeria monocytogenes Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7SCD
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-1 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7SCE
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-2 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7RSL
 
 | | Seipin forms a flexible cage at lipid droplet formation sites | | Descriptor: | Seipin | | Authors: | Arlt, H, Sui, X, Folger, B, Adams, C, Chen, X, Remme, R, Hamprecht, F.A, DiMaio, F, Liao, M, Goodman, J.M, Farese Jr, R.V, Walther, T.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Seipin forms a flexible cage at lipid droplet formation sites.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SJJ
 
 | | Crystal structure of photoactive yellow protein (PYP); F96oCNF construct | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Weaver, J.B, Kirsh, J.M, Boxer, S.G. | | Deposit date: | 2021-10-17 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Nitrile Infrared Intensities Characterize Electric Fields and Hydrogen Bonding in Protic, Aprotic, and Protein Environments.
J.Am.Chem.Soc., 144, 2022
|
|
7S6H
 
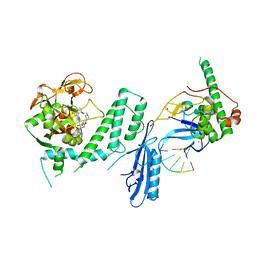 | | Human PARP1 deltaV687-E688 bound to NAD+ analog EB-47 and to a DNA double strand break. | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, DNA (5'-D(*CP*GP*AP*CP*G)-3'), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-14 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7S81
 
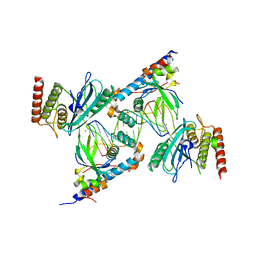 | | Structure of human PARP1 domains (Zn1, Zn3, WGR, HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*CP*GP*CP*AP*T)-3'), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7S6M
 
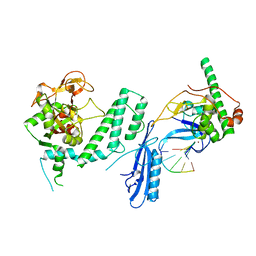 | |
