8EZD
 
 | |
8EZE
 
 | |
8F4D
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the third illumination at 2.15 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4E
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the third illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4G
 
 | | RT XFEL structure of Photosystem II 730 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4J
 
 | | RT XFEL structure of Photosystem II 4000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
1ZPH
 
 | | Crystal structure analysis of the minor groove binding quinolinium quaternary salt SN 8315 complexed with CGCGAATTCGCG | | Descriptor: | 1,6-DIMETHYL-4-(4-(4-(1-METHYLPYRIDINIUM-4-YLAMINO)PHENYLCARBAMOYL)PHENYLAMINO)QUINOLINIUM, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Adams, A, Leong, C, Denny, W.A, Guss, J.M. | | Deposit date: | 2005-05-16 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two minor-groove-binding quinolinium quaternary salts complexed with d(CGCGAATTCGCG)(2) at 1.6 and 1.8 Angstrom resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Z2Q
 
 | |
1ZUN
 
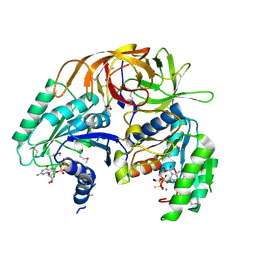 | | Crystal Structure of a GTP-Regulated ATP Sulfurylase Heterodimer from Pseudomonas syringae | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Mougous, J.D, Lee, D.H, Hubbard, S.C, Schelle, M.W, Vocadlo, D.J, Berger, J.M, Bertozzi, C.R. | | Deposit date: | 2005-05-31 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for g protein control of the prokaryotic ATP sulfurylase.
Mol.Cell, 21, 2006
|
|
1ZQ3
 
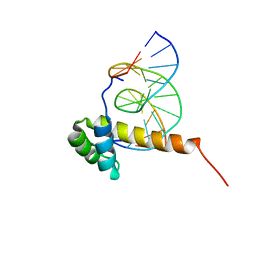 | | NMR Solution Structure of the Bicoid Homeodomain Bound to the Consensus DNA Binding Site TAATCC | | Descriptor: | 5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3', Homeotic bicoid protein | | Authors: | Baird-Titus, J.M, Rance, M, Clark-Baldwin, K, Ma, J, Vrushank, D. | | Deposit date: | 2005-05-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site.
J.Mol.Biol., 356, 2006
|
|
1ZUG
 
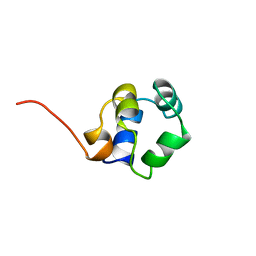 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
2BTP
 
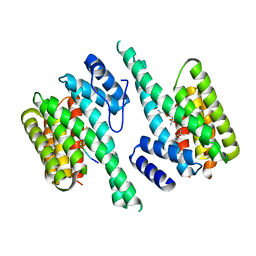 | | 14-3-3 Protein Theta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN TAU, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Johansson, A.C.E, Smee, C, Yang, X, Sundstrom, M, Edwards, A, Arrowsmith, C, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-05 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
8ERN
 
 | | Cyclin-free CDK2 in complex with Cpd21 | | Descriptor: | Cyclin-dependent kinase 2, GLYCEROL, N,3-dimethyl-4-({7-[(2S)-2-methylpyrrolidine-1-carbonyl]quinazolin-2-yl}amino)benzene-1-sulfonamide | | Authors: | Murray, J.M, Oh, A. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cyclin-free CDK2 in complex with Cpd21
To Be Published
|
|
2BCU
 
 | | DNA polymerase lambda in complex with a DNA duplex containing an unpaired Damp and a T:T mismatch | | Descriptor: | 5'-D(*CP*AP*GP*TP*TP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*AP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase
Cell(Cambridge,Mass.), 124, 2006
|
|
8ERD
 
 | | Cyclin-free CDK2 in complex with Cpd17 | | Descriptor: | (2-{[1-(methanesulfonyl)piperidin-4-yl]amino}quinazolin-7-yl)[(2S)-2-methylpyrrolidin-1-yl]methanone, Cyclin-dependent kinase 2 | | Authors: | Murray, J.M, Oh, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Cyclin-free CDK2 in complex with Cpd17
To Be Published
|
|
1Z59
 
 | | Topoisomerase VI-B, ADP-bound monomer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
1ZPI
 
 | | Crystal structure analysis of the minor groove binding quinolinium quaternary salt SN 8224 complexed with CGCGAATTCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', 8-METHOXY-1-METHYL-4-(4-(4-(1-METHYLPYRIDINIUM-4-YLAMINO)PHENYLCARBAMOYL)PHENYLAMINO)QUINOLINIUM, MAGNESIUM ION | | Authors: | Adams, A, Leong, C, Denny, W.A, Guss, J.M. | | Deposit date: | 2005-05-16 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two minor-groove-binding quinolinium quaternary salts complexed with d(CGCGAATTCGCG)(2) at 1.6 and 1.8 Angstrom resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZAX
 
 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P212121, Form B | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YYL
 
 | | crystal structure of CD4M33, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD4M33, scorpion-toxin mimic of CD4, ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
8F8K
 
 | | The structure of Rv2173 from M. tuberculosis with IPP bound | | Descriptor: | (2E,6E)-farnesyl diphosphate synthase, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ACETATE ION, ... | | Authors: | Johnston, J.M, Allison, T.M, Titterington, J, Beasley, C.P.H. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Rv2173 from M. tuberculosis in APO-, IPP-, and DMAP-bound forms.
To be Published
|
|
8F8F
 
 | | The structure of Rv2173 from M. tuberculosis (APO form) | | Descriptor: | (2E,6E)-farnesyl diphosphate synthase, GLYCEROL, MAGNESIUM ION | | Authors: | Johnston, J.M, Allison, T.M, Titterington, J. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of Rv2173 from M. tuberculosis in APO-, IPP-, and DMAP-bound forms.
To be Published
|
|
8F8L
 
 | | The structure of Rv2173 from M. tuberculosis with DMAP bound | | Descriptor: | (2E,6E)-farnesyl diphosphate synthase, CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, ... | | Authors: | Johnston, J.M, Allison, T.M, Titterington, J. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Rv2173 from M. tuberculosis in APO-, IPP-, and DMAP-bound forms.
To be Published
|
|
8FWA
 
 | | Phycocyanin structure from a modular droplet injector for serial femtosecond crystallography | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Botha, S, Doppler, D.L, Sonker, M, Egatz-Gomez, A, Grieco, A, Zaare, S, Jernigan, R, Meza-Aguilar, J.D, Rabbani, M.T, Manna, A, Alvarez, R, Karpos, K, Cruz Villarreal, J, Nelson, G, Ketawala, G.K, Pey, A.L, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Nazari, R, Sierra, R, Hunter, M.S, Batyuk, A, Kupitz, C.J, Sublett, R.E, Lisova, S, Mariani, V, Boutet, S, Fromme, R, Grant, T.D, Fromme, P, Kirian, R.A, Martin-Garcia, J.M, Ros, A. | | Deposit date: | 2023-01-20 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
8FFR
 
 | | Revised structure of the rabies virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (99-MER) | | Authors: | Leyrat, C, Bourhis, J.M, Albertini, A.A.V, Wernimont, A.K, Muziol, T, Ravelli, R.B.G, Weissenhorn, W, Ruigrok, R.W.H, Jamin, M. | | Deposit date: | 2022-12-09 | | Release date: | 2023-01-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure and Dynamics of the Unassembled Nucleoprotein of Rabies Virus in Complex with Its Phosphoprotein Chaperone Module.
Viruses, 14, 2022
|
|
1Y69
 
 | | RRF domain I in complex with the 50S ribosomal subunit from Deinococcus radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, 50S ribosomal protein L27, ... | | Authors: | Wilson, D.N, Schluenzen, F, Harms, J.M, Yoshida, T, Ohkubo, T, Albrecht, R, Buerger, J, Kobayashi, Y, Fucini, P. | | Deposit date: | 2004-12-04 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | X-ray crystallography on ribosome recycling: mechanism of binding and action of RRF on the 50S ribosomal subunit
EMBO J., 24, 2005
|
|
