1Y5L
 
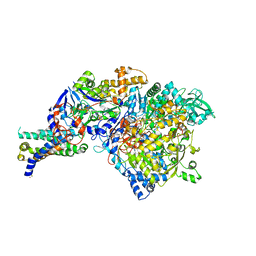 | | The crystal structure of the NarGHI mutant NarI-H66Y | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
5TY7
 
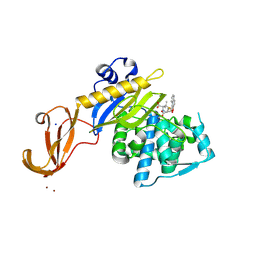 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-18 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
1XOU
 
 | |
5TX9
 
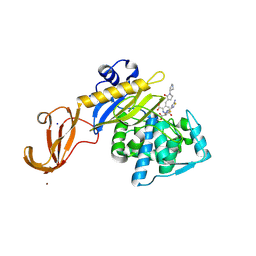 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-16 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
1Y5N
 
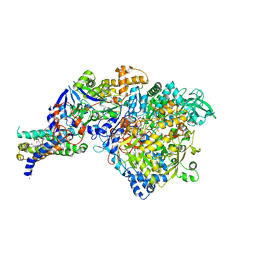 | | The crystal structure of the NarGHI mutant NarI-K86A in complex with pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5I
 
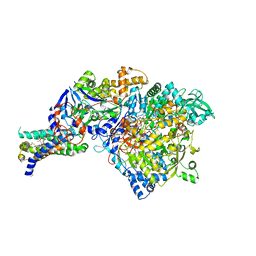 | | The crystal structure of the NarGHI mutant NarI-K86A | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
2YK4
 
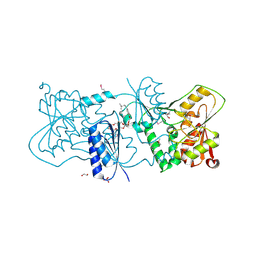 | | Structure of Neisseria LOS-specific sialyltransferase (NST). | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2YK5
 
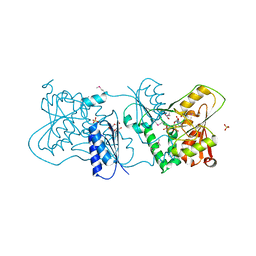 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CMP. | | Descriptor: | 1,2-ETHANEDIOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2WNB
 
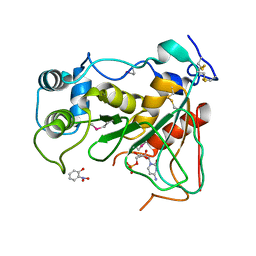 | | Crystal Structure of a Mammalian Sialyltransferase in complex with disaccharide and CMP | | Descriptor: | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase 1, CYTIDINE-5'-MONOPHOSPHATE, HYDROXY(2-HYDROXYPHENYL)OXOAMMONIUM, ... | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-08 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2WNF
 
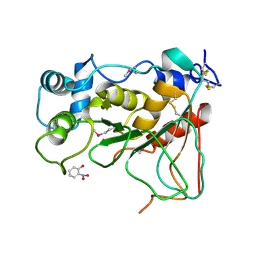 | | Crystal Structure of a Mammalian Sialyltransferase in complex with Gal-beta-1-3GalNAc-ortho-nitrophenol | | Descriptor: | CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, HYDROXY(2-HYDROXYPHENYL)OXOAMMONIUM, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2WML
 
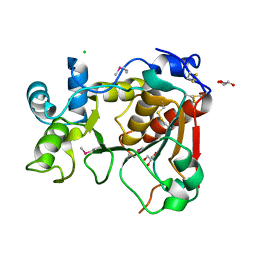 | | Crystal Structure of a Mammalian Sialyltransferase | | Descriptor: | CHLORIDE ION, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE -ALPHA-2,3-SIALYLTRANSFERASE, GLYCEROL | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-01 | | Release date: | 2009-10-13 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8CYZ
 
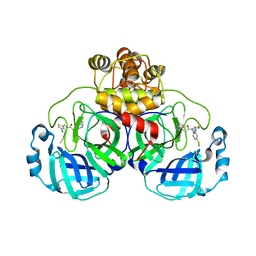 | | Crystal structure of SARS-CoV-2 Mpro with compound C4 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-[4-(methylsulfanyl)phenyl]acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ7
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C2 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-(4-methoxyphenyl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CYU
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C5 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ4
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C3 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8DRU
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp7-nsp8 (C7) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp7-nsp8 (C7) cut site, PENTAETHYLENE GLYCOL, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRV
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp8-nsp9 (C8) cut site sequence | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp8-nsp9 (C8) cut site, PENTAETHYLENE GLYCOL | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRY
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp12-nsp13 (C12) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp12-nsp13 (C12) cut site | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS0
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp14-nsp15 (C14) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRZ
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp13-nsp14 (C13) cut site sequence | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS2
 
 | | Structure of SARS-CoV-2 Mpro in complex with the nsp13-nsp14 (C13) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS1
 
 | | Structure of SARS-CoV-2 Mpro in complex with nsp12-nsp13 (C12) cut site sequence | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
2YK6
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CDP. | | Descriptor: | CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-DIPHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2WLC
 
 | | Crystallographic analysis of the polysialic acid O-acetyltransferase OatWY | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Lee, H.J, Rakic, B, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Kinetic Characterizations of the Polysialic Acid O-Acetyltransferase Oatwy from Neisseria Meningitidis.
J.Biol.Chem., 284, 2009
|
|
2WLG
 
 | | Crystallographic analysis of the polysialic acid O-acetyltransferase OatWY | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, POLYSIALIC ACID O-ACETYLTRANSFERASE, ... | | Authors: | Lee, H.J, Rakic, B, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-06-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Kinetic Characterizations of the Polysialic Acid O-Acetyltransferase Oatwy from Neisseria Meningitidis.
J.Biol.Chem., 284, 2009
|
|
