8IRC
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 5-millisecond delay (Single conformation) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8K65
 
 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase without pump laser irradiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Zoric, D, Sandelin, E, Nango, E, Tanaka, R, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
8K6Y
 
 | | Serial femtosecond crystallography structure of photo dissociated CO from ba3- type cytochrome c oxidase determined by extrapolation method | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Zoric, D, Sandelin, E, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
8IFD
 
 | | Dibekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8IFE
 
 | | Arbekacin-added human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8J6G
 
 | |
7YRU
 
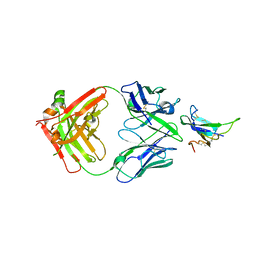 | | ALK2 antibody complex | | Descriptor: | Activin receptor type-1, antibody heavy chain, antibody light chain | | Authors: | Kawaguchi, Y, Nakamura, K, Suzuki, M, Tsuji, S, Katagiri, T. | | Deposit date: | 2022-08-10 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A blocking monoclonal antibody reveals dimerization of intracellular domains of ALK2 associated with genetic disorders.
Nat Commun, 14, 2023
|
|
6K2P
 
 | |
6K2W
 
 | |
6K2R
 
 | |
6K2V
 
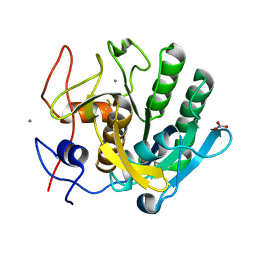 | |
6K2X
 
 | |
6K2T
 
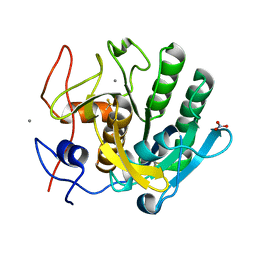 | |
6K2S
 
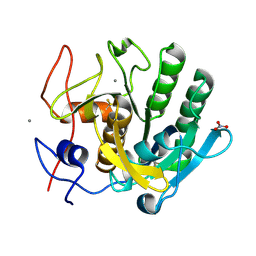 | |
8J8D
 
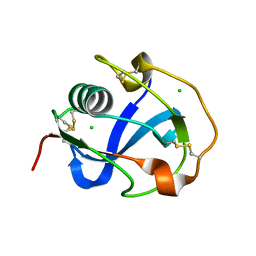 | | Crystal structure of SRCR domain 11 of DMBT1 | | Descriptor: | CHLORIDE ION, Deleted in malignant brain tumors 1 protein | | Authors: | Zhang, C, Lu, P, Nagata, K. | | Deposit date: | 2023-05-01 | | Release date: | 2023-12-13 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Refolding, Crystallization, and Crystal Structure Analysis of a Scavenger Receptor Cysteine-Rich Domain of Human Salivary Agglutinin Expressed in Escherichia coli.
Protein J., 43, 2024
|
|
8IFC
 
 | | Arbekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8IFB
 
 | | Dibekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
5ETY
 
 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | Descriptor: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | Authors: | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
6L49
 
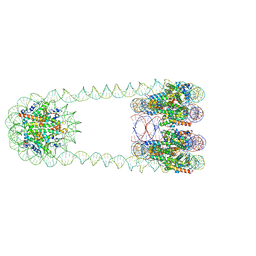 | | H3-CA-H3 tri-nucleosome with the 22 base-pair linker DNA | | Descriptor: | DNA (485-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Takizawa, Y, Ho, C.-H, Tachiwana, H, Matsunami, H, Ohi, M, Wolf, M, Kurumizaka, H. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (18.9 Å) | | Cite: | Cryo-EM Structures of Centromeric Tri-nucleosomes Containing a Central CENP-A Nucleosome.
Structure, 28, 2020
|
|
7CJI
 
 | | Photosystem II structure in the S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7CJJ
 
 | | Photosystem II structure in the S2 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7COU
 
 | | Structure of cyanobacterial photosystem II in the dark S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
3W80
 
 | |
3W7Y
 
 | |
8J4H
 
 | | X-ray structure of a ferric ion-binding protein A (FbpA) from Vibrio metschnikovii in complex with Danshensu (DSS) | | Descriptor: | (2~{R})-3-[3,4-bis(oxidanyl)phenyl]-2-oxidanyl-propanoic acid, Ferric iron ABC transporter iron-binding protein | | Authors: | Lu, P, Jiang, J, Nagata, K. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Molecular mechanism of Fe 3+ binding inhibition to Vibrio metschnikovii ferric ion-binding protein, FbpA, by rosmarinic acid and its hydrolysate, danshensu.
Protein Sci., 33, 2024
|
|
