5DFA
 
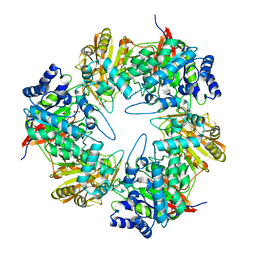 | | 3D structure of the E323A catalytic mutant of Gan42B, a GH42 beta-galactosidase from G. stearothermophilus | | Descriptor: | Beta-galactosidase, GLYCEROL, ZINC ION | | Authors: | Solomon, H.V, Tabachnikov, O, Lansky, S, Feinberg, H, Govada, L, Chayen, N.E, Shoham, Y, Shoham, G. | | Deposit date: | 2015-08-26 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function relationships in Gan42B, an intracellular GH42 beta-galactosidase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3L8Q
 
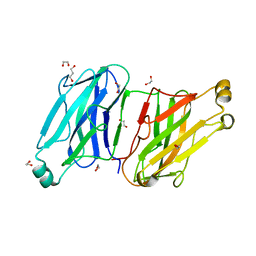 | | Structure analysis of the type II cohesin dyad from the adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2010-01-03 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Modular Arrangement of a Cellulosomal Scaffoldin Subunit Revealed from the Crystal Structure of a Cohesin Dyad
J.Mol.Biol., 399, 2010
|
|
3FNK
 
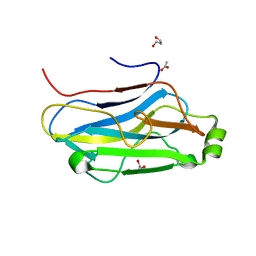 | | Crystal structure of the second type II cohesin module from the cellulosomal adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2008-12-25 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Intermodular Linker Flexibility Revealed from Crystal Structures of Adjacent Cellulosomal Cohesins of Acetivibrio cellulolyticus
J.Mol.Biol., 391, 2009
|
|
3GHP
 
 | |
8TV4
 
 | | NMR structure of temporin L in solution | | Descriptor: | Temporin-1Tl peptide | | Authors: | McShan, A.C, Jia, R, Halim, M.A. | | Deposit date: | 2023-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiviral peptides inhibiting the main protease of SARS-CoV-2 investigated by computational screening and in vitro protease assay.
J.Pept.Sci., 30, 2024
|
|
2MTE
 
 | | Solution structure of Doc48S | | Descriptor: | CALCIUM ION, Cellulose 1,4-beta-cellobiosidase (reducing end) CelS | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Revisiting the NMR solution structure of the Cel48S type-I dockerin module from Clostridium thermocellum reveals a cohesin-primed conformation.
J.Struct.Biol., 188, 2014
|
|
8A2M
 
 | |
8A2L
 
 | |
8A5N
 
 | |
6IVU
 
 | |
6IVS
 
 | |
6KG9
 
 | | Solution structure of CaDoc0917 from Clostridium acetobutylicum | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KGF
 
 | | Crystal structure of CaDoc0917(R16D)-CaCohA2 complex at pH 8.2 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
2ZF9
 
 | |
1AW5
 
 | |
1B4E
 
 | | X-ray structure of 5-aminolevulinic acid dehydratase complexed with the inhibitor levulinic acid | | Descriptor: | GLYCEROL, LAEVULINIC ACID, PROTEIN (5-AMINOLEVULINIC ACID DEHYDRATASE), ... | | Authors: | Erskine, P.T, Cooper, J.B, Lewis, G, Spencer, P, Wood, S.P, Shoolingin-Jordan, P.M. | | Deposit date: | 1998-12-19 | | Release date: | 1999-12-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 5-aminolevulinic acid dehydratase from Escherichia coli complexed with the inhibitor levulinic acid at 2.0 A resolution.
Biochemistry, 38, 1999
|
|
4P7H
 
 | |
