5XK9
 
 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with GSPP and DMAPP | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5W8I
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 23 and Zinc | | Descriptor: | 2-[3-(3,4-difluorophenyl)-5-hydroxy-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Abendroth, J. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8L
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 59 and NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-([1,1'-biphenyl]-3-yl)-5-(cyclopropylmethyl)-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, ... | | Authors: | Davies, D.R, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5XK3
 
 | | Crystal structure of apo form Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 | | Descriptor: | SULFATE ION, Undecaprenyl diphosphate synthase | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8JJC
 
 | | Tubulin-Y62 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(6,7-dimethoxy-3,4-dihydro-1~{H}-isoquinolin-2-yl)-6-(3-methoxyphenyl)pyrimidin-2-amine, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
8JJB
 
 | | Crystal structure of T2R-TTL-Y61 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
8JNX
 
 | | alkaline amylase Amy703 with truncated of N-terminus domain | | Descriptor: | Alpha-amylase, CALCIUM ION | | Authors: | Xiang, L, Zhang, G, Zhou, J. | | Deposit date: | 2023-06-06 | | Release date: | 2023-12-13 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.20279884 Å) | | Cite: | N-terminal domain truncation yielded a unique dimer of polysaccharide hydrolase with enhanced enzymatic activity, stability and calcium ion independence.
Int.J.Biol.Macromol., 266, 2024
|
|
9FKZ
 
 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[4-[(1~{R},3~{R})-3-azanylcyclopentyl]oxyphenyl]-~{N}-[(4-chlorophenyl)methyl]prop-2-enamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9FL0
 
 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[(4-chlorophenyl)methyl]-~{N}-(4-fluorophenyl)prop-2-enamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9FKY
 
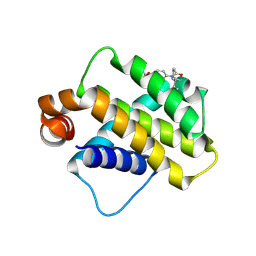 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[4-[(1~{R},3~{R})-3-azanylcyclopentyl]oxyphenyl]-~{N}-[(1~{S})-1-[3-cyano-4-(trifluoromethyl)phenyl]ethyl]propanamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
7C29
 
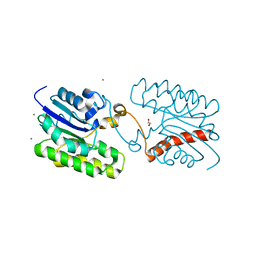 | | Esterase CrmE10 mutant-D178A | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7C82
 
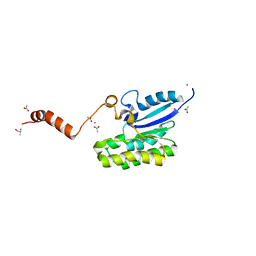 | | Crystal structure of AlinE4, a SGNH-hydrolase family esterase | | Descriptor: | ACETATE ION, ALANINE, CADMIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7C84
 
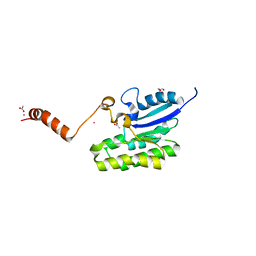 | | Esterase AlinE4 mutant, D162A | | Descriptor: | ACETATE ION, CADMIUM ION, GLYCEROL, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7C85
 
 | | Esterase AlinE4 mutant-S13A | | Descriptor: | ACETATE ION, CADMIUM ION, SGNH-hydrolase family esterase | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7DGW
 
 | | De novo designed protein H4A2S | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGU
 
 | | De novo designed protein H4A1R | | Descriptor: | de novo designed protein H4A1R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKK
 
 | | De novo design protein XM2H | | Descriptor: | De novo design protein XM2H | | Authors: | Bin, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKO
 
 | | De novo design protein AM2M | | Descriptor: | de novo designed protein AM2M | | Authors: | Bin, H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DMF
 
 | |
7DGY
 
 | | De novo designed protein H4C2R | | Descriptor: | de novo designed protein H4C2R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7C23
 
 | | Crystal structure of CrmE10, a SGNH-hydrolase family esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7Y4S
 
 | | Structure of human MG53 homo-dimer | | Descriptor: | Tripartite motif-containing protein 72 | | Authors: | Chen, L, Niu, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human MG53 homodimer.
Biochem.J., 479, 2022
|
|
7YJS
 
 | |
7YJR
 
 | |
7YJQ
 
 | | Crystal structure of MCR-1-S treated by auranofin | | Descriptor: | GOLD ION, Probable phosphatidylethanolamine transferase Mcr-1 | | Authors: | Zhang, Q, Sun, H, Wang, M. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gold drugs as colistin adjuvants in the fight against MCR-1 producing bacteria.
J.Biol.Inorg.Chem., 28, 2023
|
|
