4EEK
 
 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) With bound phosphate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, PHOSPHATE ION, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4GM6
 
 | | Crystal structure of PfkB family carbohydrate kinase(TARGET EFI-502146 FROM Listeria grayi DSM 20601 | | Descriptor: | CHLORIDE ION, GLYCEROL, PfkB family carbohydrate kinase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PfkB family carbohydrate kinase FROM Listeria grayi
To be Published
|
|
4F72
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from Bacteroides thetaiotaomicron, asp12ala mutant, complex with magnesium and inorganic phosphate | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Farelli, J.D, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Zencheck, W.D, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4F71
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from Bacteroides thetaiotaomicron, wild-type protein, complex with magnesium and inorganic phosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4HZ1
 
 | | Crystal Structure of Pseudomonas aeruginosa azurin with iron(II) at the copper-binding site. | | Descriptor: | ACETATE ION, Azurin, FE (II) ION | | Authors: | McLaughlin, M.P, Retegan, M, Bill, E, Payne, T.M, Shafaat, H.S, Pea, S, Sudhamsu, J, Ensign, A.A, Crane, B.R, Neese, F, Holland, P.L. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azurin as a Protein Scaffold for a Low-coordinate Nonheme Iron Site with a Small-molecule Binding Pocket.
J.Am.Chem.Soc., 134, 2012
|
|
2C52
 
 | | Structural diversity in CBP p160 complexes | | Descriptor: | CREB-BINDING PROTEIN, NUCLEAR RECEPTOR COACTIVATOR 1 | | Authors: | Waters, L.C, Yue, B, Veverka, V, Renshaw, P.S, Bramham, J, Matsuda, S, Frenkiel, T, Kelly, G, Muskett, F.W, Carr, M.D, Heery, D.M. | | Deposit date: | 2005-10-25 | | Release date: | 2006-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in p160/CREB-binding protein coactivator complexes.
J. Biol. Chem., 281, 2006
|
|
2BXT
 
 | | Design and Discovery of Novel, Potent Thrombin Inhibitors with a Solubilizing Cationic P1-P2-Linker | | Descriptor: | 6-CHLORO-1-(2-{[(5-CHLORO-1-BENZOTHIEN-3-YL)METHYL]AMINO}ETHYL)-3-[(2-PYRIDIN-2-YLETHYL)AMINO]-1,4-DIHYDROPYRAZIN-2-OL, ALPHA THROMBIN, HIRUDIN VARIANT-2 | | Authors: | Bulat, S, Bosio, S, Grabowski, E, Papadopoulos, M.A, Cerezo-Galvez, S, Rosenbaum, C, Matassa, V.G, Ott, I, Metz, G, Schamberger, J, Sekul, R, Feurer, A. | | Deposit date: | 2005-07-27 | | Release date: | 2006-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and Discovery of Novel, Potent Pyrazinone-Based Thrombin Inhibitors with a Solubilizing P1-P2-Linker
Lett.Drug Des.Discovery, 3, 2006
|
|
1ONV
 
 | | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNAP II CTD Phosphatase FCP1 | | Descriptor: | Transcription initiation factor IIF, alpha subunit, serine phosphatase FCP1a | | Authors: | Nguyen, B.D, Abbott, K.L, Potempa, K, Kobor, M.S, Archambault, J, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNA polymerase II carboxyl-terminal domain phosphatase FCP1
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2BVX
 
 | | Design and Discovery of Novel, Potent Thrombin Inhibitors with a Solubilizing Cationic P1-P2-Linker | | Descriptor: | ALPHA THROMBIN, HIRUDIN VARIANT-2, N-(5-CHLORO-BENZO[B]THIOPHEN-3-YLMETHYL)-2-[6-CHLORO-OXO-3-(2-PYRIDIN-2-YL-ETHYLAMINO)-2H-PYRAZIN-1-YL]-ACETAMIDE | | Authors: | Bulat, S, Bosio, S, Grabowski, E, Papadopoulos, M.A, Cerezo-Galvez, S, Rosenbaum, C, Matassa, V.G, Ott, I, Metz, G, Schamberger, J, Sekul, R, Feurer, A. | | Deposit date: | 2005-07-04 | | Release date: | 2006-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and Discovery of Novel, Potent Pyrazinone-Based Thrombin Inhibitors with a Solubilizing P1-P2-Linker
Lett.Drug Des.Discovery, 3, 2006
|
|
2BXU
 
 | | Design and Discovery of Novel, Potent Thrombin Inhibitors with a Solubilizing Cationic P1-P2-Linker | | Descriptor: | 1-(2-{[(6-AMINO-2-METHYLPYRIDIN-3-YL)METHYL]AMINO}ETHYL)-6-CHLORO-3-[(2,2-DIFLUORO-2-PYRIDIN-2-YLETHYL)AMINO]-1,4-DIHYDROPYRAZIN-2-OL, ALPHA THROMBIN, HIRUDIN | | Authors: | Bulat, S, Bosio, S, Grabowski, E, Papadopoulos, M.A, Cerezo-Galvez, S, Rosenbaum, C, Matassa, V.G, Ott, I, Metz, G, Schamberger, J, Sekul, R, Feurer, A. | | Deposit date: | 2005-07-27 | | Release date: | 2006-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Discovery of Novel, Potent Pyrazinone-Based Thrombin Inhibitors with a Solubilizing P1-P2-Linker
Lett.Drug Des.Discovery, 3, 2006
|
|
111D
 
 | |
2ROG
 
 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
1JWI
 
 | | Crystal Structure of Bitiscetin, a von Willeband Factor-dependent Platelet Aggregation Inducer. | | Descriptor: | bitiscetin, platelet aggregation inducer | | Authors: | Hirotsu, S, Mizuno, H, Fukuda, K, Qi, M.C, Matsui, T, Hamako, J, Morita, T, Titani, K. | | Deposit date: | 2001-09-04 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of bitiscetin, a von Willebrand factor-dependent platelet aggregation inducer.
Biochemistry, 40, 2001
|
|
1SBS
 
 | | CRYSTAL STRUCTURE OF AN ANTI-HCG FAB | | Descriptor: | MONOCLONAL ANTIBODY 3A2, SULFATE ION | | Authors: | Fotinou, C, Beauchamp, J, Emsley, P, Dehaan, A, Schielen, W.J.G, Bos, E, Isaacs, N.W. | | Deposit date: | 1998-04-08 | | Release date: | 1999-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an Fab fragment against a C-terminal peptide of hCG at 2.0 A resolution.
J.Biol.Chem., 273, 1998
|
|
8VGH
 
 | |
8VGL
 
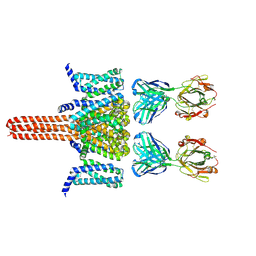 | | CryoEM structure of Nav1.7 in complex with wild type Fab 7A9 | | Descriptor: | Chimeric Nav1.7-NavAb, Fab 7A9 heavy chain, Fab 7A9 light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Disulfi de constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
8VEG
 
 | |
8VGF
 
 | |
8VGG
 
 | |
8VGE
 
 | |
8VGM
 
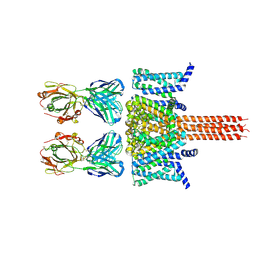 | | CryoEM structure of Nav1.7 in complex with engineered conformationally rigid Fab 7A9.4DS | | Descriptor: | Chimeric Nav1.7-NavAb, Fab 7A9.4DS heavy chain, Fab 7A9.4DS light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Disulfi de constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
8VGQ
 
 | |
8VGJ
 
 | |
8VGI
 
 | |
