4BZZ
 
 | | Complete crystal structure of carboxylesterase Cest-2923 from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, ACETONITRILE, LIPASE/ESTERASE, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, de las Rivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
4C08
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with CaCl2 at 1.34 Angstroms | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.338 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
3BUT
 
 | | Crystal structure of protein Af_0446 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein Af_0446 | | Authors: | Bonanno, J.B, Patskovsky, Y, Ozyurt, S, Ashok, S, Zhang, F, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-03 | | Release date: | 2008-01-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of protein Af_0446 from Archaeoglobus fulgidus.
To be Published
|
|
1AFS
 
 | | RECOMBINANT RAT LIVER 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE (3-ALPHA-HSD) COMPLEXED WITH NADP AND TESTOSTERONE | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TESTOSTERONE | | Authors: | Bennett, M.J, Albert, R.H, Jez, J.M, Ma, H, Penning, T.M, Lewis, M. | | Deposit date: | 1997-03-13 | | Release date: | 1997-10-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
Structure, 5, 1997
|
|
4BQS
 
 | | Crystal structure of Mycobacterium tuberculosis shikimate kinase in complex with ADP and a shikimic acid derivative. | | Descriptor: | (1R,6R,10S)-6,10-dihydroxy-2-oxabicyclo[4.3.1]deca-4(Z),7-diene-8-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, SHIKIMATE KINASE | | Authors: | Otero, J.M, Garcia-Doval, C, Llamas-Saiz, A.L, Blanco, B, Prado, V, Lence, E, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2013-06-02 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mycobacterium tuberculosis shikimate kinase inhibitors: design and simulation studies of the catalytic turnover.
J. Am. Chem. Soc., 135, 2013
|
|
1ZPH
 
 | | Crystal structure analysis of the minor groove binding quinolinium quaternary salt SN 8315 complexed with CGCGAATTCGCG | | Descriptor: | 1,6-DIMETHYL-4-(4-(4-(1-METHYLPYRIDINIUM-4-YLAMINO)PHENYLCARBAMOYL)PHENYLAMINO)QUINOLINIUM, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Adams, A, Leong, C, Denny, W.A, Guss, J.M. | | Deposit date: | 2005-05-16 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two minor-groove-binding quinolinium quaternary salts complexed with d(CGCGAATTCGCG)(2) at 1.6 and 1.8 Angstrom resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6FVX
 
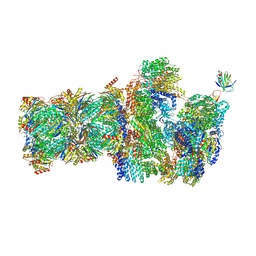 | | 26S proteasome, s5 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
4C06
 
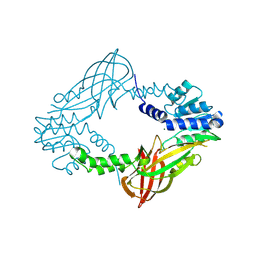 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with MgCl2 | | Descriptor: | MAGNESIUM ION, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
4C05
 
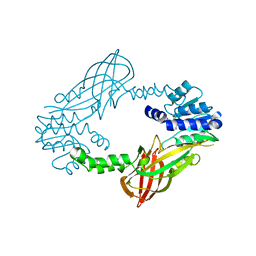 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with SAH | | Descriptor: | PROTEIN ARGININE N-METHYLTRANSFERASE 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
1Z2Q
 
 | |
3BU1
 
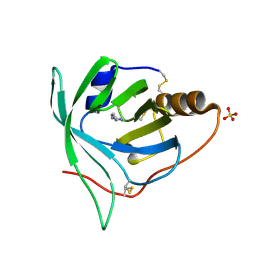 | | Crystal structure of monomine-histamine complex | | Descriptor: | HISTAMINE, Lipocalin, SULFATE ION | | Authors: | Mans, B.J, Ribeiro, J.M, Andersen, J.F. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, function, and evolution of biogenic amine-binding proteins in soft ticks.
J.Biol.Chem., 283, 2008
|
|
1Y4Y
 
 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
6GNY
 
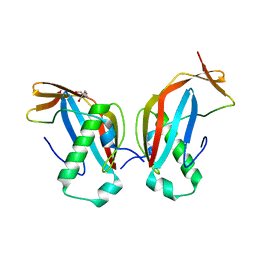 | | Crystal structure of the MAJIN-TERB2 heterotetrameric complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Membrane-anchored junction protein, Telomere repeats-binding bouquet formation protein 2 | | Authors: | Dunce, J.M, Gurusaran, M, Sen, L.T, Davies, O.R. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of meiotic telomere attachment to the nuclear envelope by MAJIN-TERB2-TERB1.
Nat Commun, 9, 2018
|
|
6GRA
 
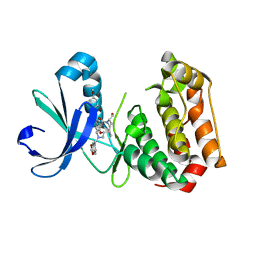 | | Human AURKA bound to BRD-7880 | | Descriptor: | 1-[(2~{R},3~{S})-2-[[1,3-benzodioxol-5-ylmethyl(methyl)amino]methyl]-3-methyl-6-oxidanylidene-5-[(2~{S})-1-oxidanylpropan-2-yl]-3,4-dihydro-2~{H}-1,5-benzoxazocin-8-yl]-3-(4-methoxyphenyl)urea, Aurora kinase A | | Authors: | Abdul Azeez, K.R, Sorrell, F.J, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aurora kinase A bound to BRD-7880
To Be Published
|
|
3C3K
 
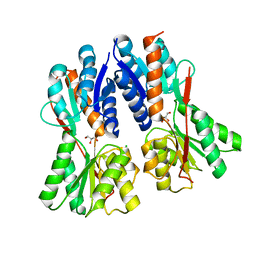 | | Crystal structure of an uncharacterized protein from Actinobacillus succinogenes | | Descriptor: | Alanine racemase, CHLORIDE ION, GLYCEROL | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of an uncharacterized protein from Actinobacillus succinogenes.
To be Published
|
|
4F0R
 
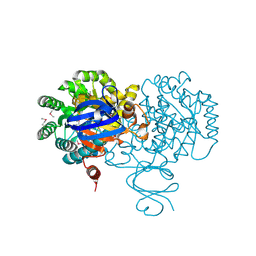 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-methylthioadenosine/S-adenosylhomocysteine deaminase, GLYCEROL, ... | | Authors: | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-06-06 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex)
To be Published
|
|
3B5M
 
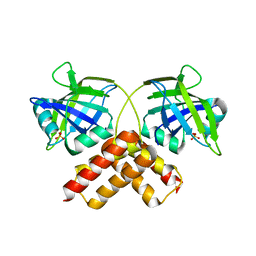 | | Crystal structure of conserved uncharacterized protein from Rhodopirellula baltica | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Bonanno, J.B, Sridhar, V, Rutter, M, Powell, A, Maletic, M, Rodgers, R, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal Structure of Conserved Uncharacterized Protein from Rhodopirellula baltica.
To be Published
|
|
3B6B
 
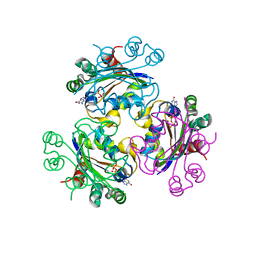 | | Crystal structure of Acanthamoeba polyphaga mimivirus nucleoside diphosphate kinase complexed with dGDP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2007-10-28 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
1Y50
 
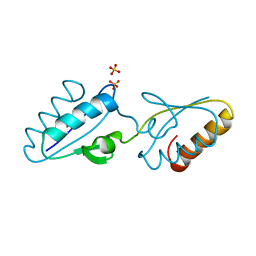 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) F29W mutant domain_swapped dimer | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
1YH5
 
 | | Solution NMR Structure of Protein yggU from Escherichia coli. Northeast Structural Genomics Consortium Target ER14. | | Descriptor: | ORF, HYPOTHETICAL PROTEIN | | Authors: | Aramini, J.M, Xiao, R, Huang, Y.J, Acton, T.B, Wu, M.J, Mills, J.L, Tejero, R.T, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Hypothetical Protein Yggu from E. Coli. Northeast Structural Genomics Consortium Target Er14.
To be Published
|
|
3B1X
 
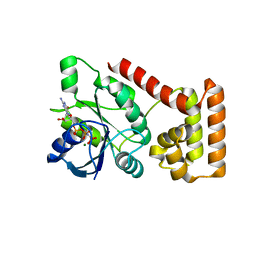 | | Crystal structure of an S. thermophilus NFeoB E66A mutant bound to GMPPNP | | Descriptor: | Ferrous iron uptake transporter protein B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-15 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A suite of Switch I and Switch II mutant structures from the G-protein domain of FeoB
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6G4A
 
 | | FLN5 (full length) | | Descriptor: | Gelation factor | | Authors: | Waudby, C.A, Wlodarski, T, Karyadi, M.-E, Cassaignau, A.M.E, Chan, S.H.S, Wentink, A.S, Schmidt-Engler, J.M, Camilloni, C, Vendruscolo, M, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mapping energy landscapes of a growing filamin domain reveals an intermediate associated with proline isomerization during biosynthesis
To Be Published
|
|
1ZPI
 
 | | Crystal structure analysis of the minor groove binding quinolinium quaternary salt SN 8224 complexed with CGCGAATTCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', 8-METHOXY-1-METHYL-4-(4-(4-(1-METHYLPYRIDINIUM-4-YLAMINO)PHENYLCARBAMOYL)PHENYLAMINO)QUINOLINIUM, MAGNESIUM ION | | Authors: | Adams, A, Leong, C, Denny, W.A, Guss, J.M. | | Deposit date: | 2005-05-16 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two minor-groove-binding quinolinium quaternary salts complexed with d(CGCGAATTCGCG)(2) at 1.6 and 1.8 Angstrom resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YYL
 
 | | crystal structure of CD4M33, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD4M33, scorpion-toxin mimic of CD4, ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
4E0U
 
 | | Crystal structure of CdpNPT in complex with thiolodiphosphate and (S)-benzodiazependione | | Descriptor: | (3S)-3-(1H-indol-3-ylmethyl)-3,4-dihydro-1H-1,4-benzodiazepine-2,5-dione, 1,2-ETHANEDIOL, Cyclic dipeptide N-prenyltransferase, ... | | Authors: | Schuller, J.M, Zocher, G, Stehle, T. | | Deposit date: | 2012-03-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity.
J.Mol.Biol., 422, 2012
|
|
