2KQT
 
 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | Authors: | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
1QRZ
 
 | | CATALYTIC DOMAIN OF PLASMINOGEN | | Descriptor: | PLASMINOGEN | | Authors: | Peisach, E, Wang, J, de los Santos, T, Reich, E, Ringe, D. | | Deposit date: | 1999-06-16 | | Release date: | 1999-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the proenzyme domain of plasminogen.
Biochemistry, 38, 1999
|
|
2DK9
 
 | | Solution structure of Calponin Homology domain of Human MICAL-1 | | Descriptor: | NEDD9-interacting protein with calponin homology and LIM domains | | Authors: | Sun, H, Dai, H, Zhang, J, Xiong, S, Wu, J, Shi, Y. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calponin homology domain of Human MICAL-1
J.Biomol.Nmr, 36, 2006
|
|
6A4U
 
 | | The first crystal structure of crustacean ferritin that is a hybrid type of H and L ferritin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ferritin, ... | | Authors: | Masuda, T, Mikami, B, Zang, J, Zhao, G. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The first crystal structure of crustacean ferritin that is a hybrid type of H and L ferritin
Protein Sci., 27, 2018
|
|
3S7X
 
 | | Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1, ... | | Authors: | Neu, U, Wang, J, Stehle, T. | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the major capsid proteins of the human KI and WU polyomaviruses.
J.Virol., 85, 2011
|
|
7W80
 
 | | Crystal Structure of the Heterodimeric HIF-2 in Complex with Antagonist Belzutifan | | Descriptor: | 3-{[(1S,2S,3R)-2,3-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Ren, X, Diao, X, Zhuang, J, Wu, D. | | Deposit date: | 2021-12-07 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.754 Å) | | Cite: | Structural basis for the allosteric inhibition of hypoxia-inducible factor (HIF)-2 by belzutifan.
Mol.Pharmacol., 2022
|
|
3RWU
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dATP Opposite Difluorotoluene Nucleoside | | Descriptor: | 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3', 5'-D(*TP*CP*GP*(DFT)P*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-05-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Hydrogen-Bonding Capability of a Templating Difluorotoluene Nucleotide Residue in an RB69 DNA Polymerase Ternary Complex.
J.Am.Chem.Soc., 133, 2011
|
|
7CS1
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Neomycin | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, NEOMYCIN | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CSI
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside 2'-N-acetyltransferase, COENZYME A | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CSJ
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Gentamicin | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, gentamicin C1 | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CRM
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-APO Structure | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside 2'-N-acetyltransferase | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CS0
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Paromomycin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
2LAC
 
 | | NMR structure of unmodified_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
2Y7J
 
 | | Structure of human phosphorylase kinase, gamma 2 | | Descriptor: | N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, PHOSPHORYLASE B KINASE GAMMA CATALYTIC CHAIN, TESTIS/LIVER ISOFORM | | Authors: | Muniz, J.R.C, Shrestha, A, Savitsky, P, Wang, J, Rellos, P, Fedorov, O, Burgess-Brown, N, Brenner, B, Berridge, G, Elkins, J.M, Krojer, T, Vollmar, M, Che, K.H, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Phosphorylase Kinase, Gamma 2
To be Published
|
|
2LJ8
 
 | |
3SIP
 
 | | Crystal structure of drICE and dIAP1-BIR1 complex | | Descriptor: | Apoptosis 1 inhibitor, Caspase, ZINC ION | | Authors: | Li, X, Wang, J, Shi, Y. | | Deposit date: | 2011-06-20 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.496 Å) | | Cite: | Structural mechanisms of DIAP1 auto-inhibition and DIAP1-mediated inhibition of drICE.
Nat Commun, 2, 2011
|
|
2LBR
 
 | |
7CVH
 
 | | Human Fructose-1,6-bisphosphatase 1 in complex with geranylgeranyl diphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structural basis for GGPP activation on the enzymatic activities FBP1
To Be Published
|
|
3RZC
 
 | | Structure of the self-antigen iGb3 bound to mouse CD1d and in complex with the iNKT TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, Beta-2-microglobulin, ... | | Authors: | Yu, E.D, Girardi, E, Wang, J, Zajonc, D.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cutting Edge: Structural Basis for the Recognition of {beta}-Linked Glycolipid Antigens by Invariant NKT Cells.
J.Immunol., 187, 2011
|
|
2VPH
 
 | | Crystal structure of the human protein tyrosine phosphatase, non- receptor type 4, PDZ domain | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 4 | | Authors: | Roos, A.K, Wang, J, Burgess-Brown, N, Elkins, J.M, Kavanagh, K, Pike, A.C.W, Filippakopoulos, P, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Bountra, C, Knapp, S. | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Protein Tyrosine Phosphatase, Non-Receptor Type 4, Pdz Domain
To be Published
|
|
7D5Y
 
 | |
2WNT
 
 | | Crystal Structure of the Human Ribosomal protein S6 kinase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, RIBOSOMAL PROTEIN S6 KINASE, ... | | Authors: | Muniz, J.R.C, Elkins, J.M, Wang, J, Ugochukwu, E, Salah, E, King, O, Picaud, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Ribosomal Protein S6 Kinase
To be Published
|
|
2LBQ
 
 | | NMR structure of i6A37_tyrASL | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*(6IA)P*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-04-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
2EFG
 
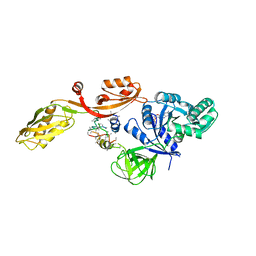 | | TRANSLATIONAL ELONGATION FACTOR G COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PROTEIN (ELONGATION FACTOR G DOMAIN 3), PROTEIN (ELONGATION FACTOR G) | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1998-09-23 | | Release date: | 1999-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
2AOR
 
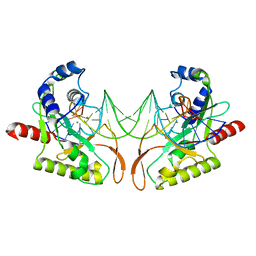 | | Crystal structure of MutH-hemimethylated DNA complex | | Descriptor: | 5'-D(*CP*AP*GP*GP*(6MA)P*TP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*AP*TP*CP*CP*TP*G)-3', CALCIUM ION, DNA mismatch repair protein mutH | | Authors: | Lee, J.Y, Chang, J, Joseph, N, Ghirlando, R, Rao, D.N, Yang, W. | | Deposit date: | 2005-08-13 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage.
Mol.Cell, 20, 2005
|
|
