1HTT
 
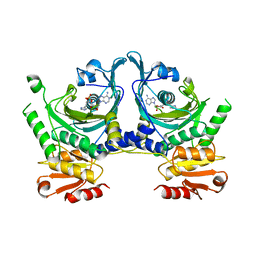 | | HISTIDYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, HISTIDINE, HISTIDYL-TRNA SYNTHETASE | | Authors: | Arnez, J.G, Harris, D.C, Mitschler, A, Rees, B, Francklyn, C.S, Moras, D. | | Deposit date: | 1996-03-09 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate.
EMBO J., 14, 1995
|
|
5QOI
 
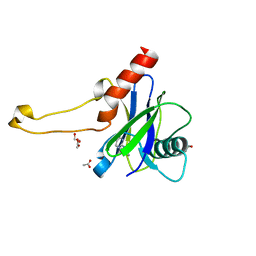 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000213a | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7NH3
 
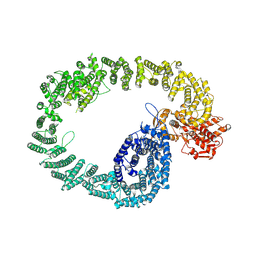 | | Nematocida Huwe1 in open conformation. | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Petrova, O, Grishkovskaya, I, Grabarczyk, D.B, Kessler, D, Haselbach, D, Clausen, T. | | Deposit date: | 2021-02-09 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.37 Å) | | Cite: | Crystal structure of HUWE1: One ring to ubiquitinate them all
To Be Published
|
|
5QP9
 
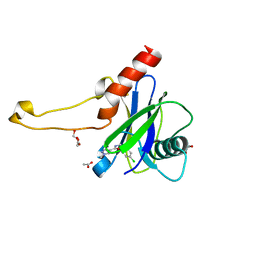 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z100435060 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
8XXS
 
 | | Crystal structure of PDE4D catalytic domain complexed with L11 | | Descriptor: | 2-[4-[bis(fluoranyl)methoxy]-3-(cyclopropylmethoxy)phenyl]-1-benzofuran-6-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.10000944 Å) | | Cite: | 5-hydroxymethylcytosine features of portal venous blood predict metachronous liver metastases of colorectal cancer and reveal phosphodiesterase 4 as a therapeutic target.
Clin Transl Med, 15, 2025
|
|
9CYB
 
 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P1 | | Descriptor: | Papain-like protease, SUCCINIC ACID, [(3R)-1-cyclopentylpiperidin-3-yl](6-methoxynaphthalen-2-yl)methanone | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYD
 
 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P4 | | Descriptor: | (1S,4s)-4-{(3R)-3-[(E)-(methoxyimino)(6-methoxynaphthalen-2-yl)methyl]piperidin-1-yl}cyclohexan-1-ol, CHLORIDE ION, Papain-like protease, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYK
 
 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P24 | | Descriptor: | ACETIC ACID, GLYCEROL, Papain-like protease, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
5QOS
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1187701032 | | Descriptor: | (3S)-N-(pyrimidin-2-yl)azepan-3-amine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
9CYC
 
 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P2 | | Descriptor: | (E)-1-[(3R)-1-cyclopentylpiperidin-3-yl]-N-methoxy-1-(6-methoxynaphthalen-2-yl)methanimine, ACETIC ACID, GLYCEROL, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchel, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
8YA2
 
 | | Structure of the SecA-SecY complex with the substrate FtsQ-LacY(+20C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division protein FtsQ,Lactose permease, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8YA0
 
 | | Structure of the SecA-SecY complex with the substrate FtsQ-LacY(+7C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division protein FtsQ,Lactose permease, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8YA3
 
 | | Structure of the SecA-SecY complex with the substrate FtsQ-LacY(+7C) treated with DTT | | Descriptor: | Cell division protein FtsQ,Lactose permease, Protein translocase subunit SecE, Protein translocase subunit SecY | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8YAS
 
 | | Structure of the SecA-SecY complex with the substrate HmBRI-7TM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-10 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8Y9Z
 
 | | Structure of the SecA-SecY complex with the substrate HmBRI-3TM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Bacteriorhodopsin-I, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8Y9Y
 
 | | Structure of the SecA-SecY complex with the substrate FtsQ-LacY(+1C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
5QOJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOCR000171b | | Descriptor: | 1,2-ETHANEDIOL, 2-(thiophen-2-yl)-1H-imidazole, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QP3
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1494850193 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6V05
 
 | | Cryo-EM structure of a substrate-engaged Bam complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
5KNZ
 
 | | Human Islet Amyloid Polypeptide Segment 19-SGNNFGAILSS-29 with Early Onset S20G Mutation Determined by MicroED | | Descriptor: | hIAPP(residues 19-29)S20G | | Authors: | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7OK0
 
 | | Cryo-EM structure of the Sulfolobus acidocaldarius RNA polymerase at 2.88 A | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-05-17 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OQY
 
 | | Cryo-EM structure of the cellular negative regulator TFS4 bound to the archaeal RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-04 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OQ4
 
 | | Cryo-EM structure of the ATV RNAP Inhibitory Protein (RIP) bound to the DNA-binding channel of the host's RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-02 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
