5HWI
 
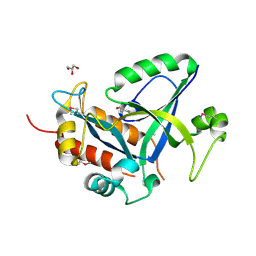 | | Crystal structure of selenomethionine labelled gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | GLYCEROL, Glutathione-specific gamma-glutamylcyclotransferase, SUCCINIC ACID | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
5HWK
 
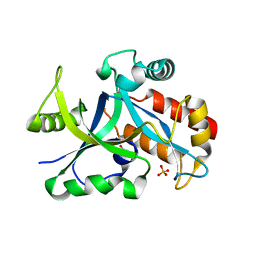 | | Crystal structure of gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | BENZOIC ACID, Glutathione-specific gamma-glutamylcyclotransferase, PHOSPHATE ION | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.344 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
3FS3
 
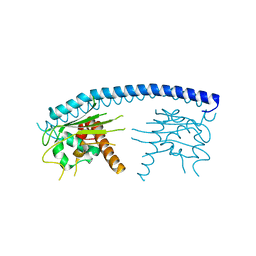 | | Crystal structure of malaria parasite Nucleosome Assembly Protein (NAP) | | Descriptor: | Nucleosome assembly protein 1, putative | | Authors: | Gill, J, Yogavel, M, Kumar, A, Belrhali, H, Sharma, A. | | Deposit date: | 2009-01-09 | | Release date: | 2009-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of malaria parasite nucleosome assembly protein: distinct modes of protein localization and histone recognition.
J.Biol.Chem., 284, 2009
|
|
8XZC
 
 | |
5IQY
 
 | | Structure of apo-Dehydroascorbate Reductase from Pennisetum Glaucum phased by Iodide-SAD method | | Descriptor: | Dehydroascorbate reductase, IODIDE ION | | Authors: | Das, B.K, Kumar, A, Manidola, P, Arockiasamy, A. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase
Biochem.Biophys.Res.Commun., 473, 2016
|
|
7XBJ
 
 | | Txp40, an insecticidal toxin protein from Xenorhabdus nematophila | | Descriptor: | 40kDa insecticidal toxin | | Authors: | Kinkar, O, Kumar, A, Prashar, A, Hire, R.S, Makde, R.D. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The crystal structure of insecticidal protein Txp40 from Xenorhabdus nematophila reveals a two-domain unique binary toxin with homology to the toxin-antitoxin (TA) system.
Insect Biochem.Mol.Biol., 164, 2023
|
|
7VMH
 
 | |
7VMI
 
 | |
8J9C
 
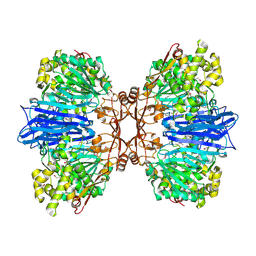 | | Crystal structure of M61 peptidase (apo-form) from Xanthomonas campestris | | Descriptor: | GLYCEROL, Putative glycyl aminopeptidase, SODIUM ION, ... | | Authors: | Yadav, P, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 291, 2024
|
|
8J9D
 
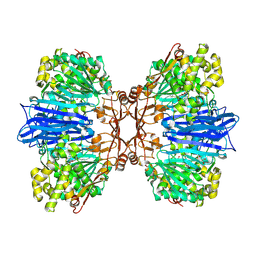 | | Crystal structure of M61 peptidase (bestatin-bound) from Xanthomonas campestris | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yadav, P, Kumar, A, Kulkarni, B.S, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 291, 2024
|
|
8JFS
 
 | | Phosphate bound acylphosphatase from Deinococcus radiodurans at 1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Acylphosphatase, CITRIC ACID, ... | | Authors: | Khakerwala, Z, Kumar, A, Makde, R.D. | | Deposit date: | 2023-05-18 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of phosphate bound Acyl phosphatase mini-enzyme from Deinococcus radiodurans at 1 angstrom resolution.
Biochem.Biophys.Res.Commun., 671, 2023
|
|
4B2Z
 
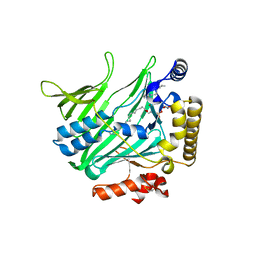 | | Structure of Osh6 in complex with phosphatidylserine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Maeda, K, Anand, K, Chiapparino, A, Kumar, A, Poletto, M, Kaksonen, M, Gavin, A.C. | | Deposit date: | 2012-07-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interactome Map Uncovers Phosphatidylserine Transport by Oxysterol-Binding Proteins
Nature, 501, 2013
|
|
7D5L
 
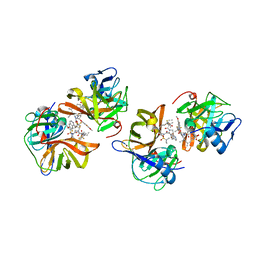 | | Discovery of BMS-986144, a Third Generation, Pan Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection | | Descriptor: | NS3/4A Protease, ZINC ION, [1,1,1-tris(fluoranyl)-2-methyl-propan-2-yl] ~{N}-[(1~{S},4~{R},6~{S},7~{Z},11~{R},13~{R},14~{S},18~{R})-13-ethyl-18-(7-fluoranyl-6-methoxy-isoquinolin-1-yl)oxy-11-methyl-4-[(1-methylcyclopropyl)sulfonylcarbamoyl]-2,15-bis(oxidanylidene)-3,16-diazatricyclo[14.3.0.0^{4,6}]nonadec-7-en-14-yl]carbamate | | Authors: | Ghosh, K, Anumula, R, Kumar, A. | | Deposit date: | 2020-09-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986144, a Third-Generation, Pan-Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 63, 2020
|
|
8OUI
 
 | | Complex of ASCT2 with Suppressyn | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Suppressyn | | Authors: | Khare, S, Kumar, A, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6EOJ
 
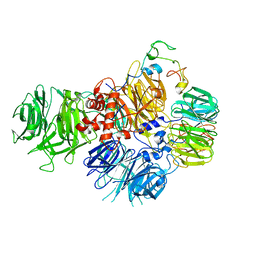 | | PolyA polymerase module of the cleavage and polyadenylation factor (CPF) from Saccharomyces cerevisiae | | Descriptor: | Polyadenylation factor subunit 2,Polyadenylation factor subunit 2, Protein CFT1, ZINC ION, ... | | Authors: | Casanal, A, Kumar, A, Hill, C.H, Emsley, P, Passmore, L.A. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Architecture of eukaryotic mRNA 3'-end processing machinery.
Science, 358, 2017
|
|
7BB4
 
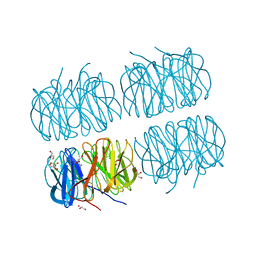 | | Crystal structure of perdeuterated PLL lectin in complex with L-fucose | | Descriptor: | GLYCEROL, PLL lectin, alpha-L-fucopyranose, ... | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7BBI
 
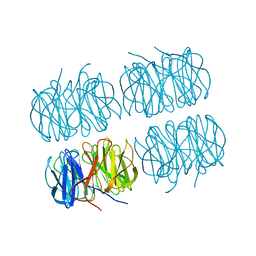 | | Joint X-ray/neutron room temperature structure of H/D-exchanged PLL lectin | | Descriptor: | PLL lectin | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-17 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7BBC
 
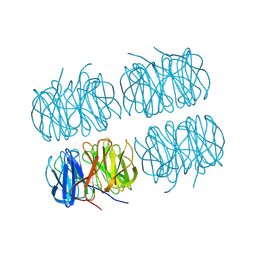 | | Joint X-ray/neutron room temperature structure of perdeuterated PLL lectin in complex with perdeuterated L-fucose | | Descriptor: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-17 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-09 | | Method: | NEUTRON DIFFRACTION (1.84 Å), X-RAY DIFFRACTION | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7QV7
 
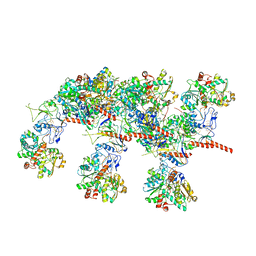 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | Descriptor: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | Authors: | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | Deposit date: | 2022-01-19 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
8V38
 
 | |
4FNO
 
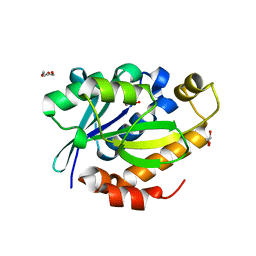 | | Crystal structure of peptidyl t-RNA hydrolase from Pseudomonas aeruginosa at 2.2 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Arora, A, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
5GIU
 
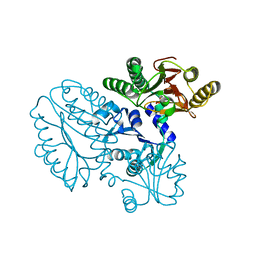 | | Crystal structure of Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, SODIUM ION | | Authors: | Are, V.N, Kumar, A, Singh, R, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site
To Be Published
|
|
5GJL
 
 | | Solution structure of SUMO from Plasmodium falciparum | | Descriptor: | Uncharacterized protein | | Authors: | Singh, J.S, Shukla, V.K, Gujrati, M, Mishra, R.K, Kumar, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interaction study of SUMO from Plasmodium falciparum
To Be Published
|
|
5I4F
 
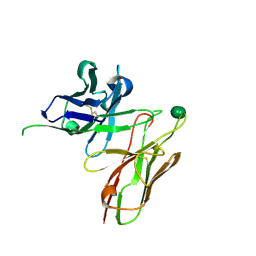 | | scFv 2D10 complexed with alpha 1,6 mannobiose | | Descriptor: | alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, scFv 2D10 | | Authors: | Vashisht, S, Kumar, A, Kaur, K.J, Salunke, D.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Antibodies Can Exploit Molecular Crowding to Bind New Antigens at Noncanonical Paratope Positions
CHEMISTRYSELECT, 1, 2016
|
|
7TJI
 
 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with flexible Orc6 N-terminal domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | Authors: | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | Deposit date: | 2022-01-16 | | Release date: | 2022-10-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
