2EVV
 
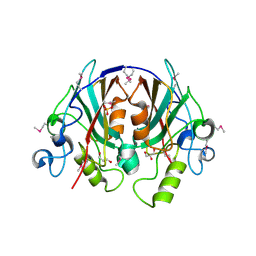 | | Crystal Structure of the PEBP-like Protein of Unknown Function HP0218 from Helicobacter pylori | | Descriptor: | GLYCEROL, SULFATE ION, hypothetical protein HP0218 | | Authors: | Kim, Y, Xu, X, Hong, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of the Hypothetical Protein HP0218 from Helicobacter pylori
To be Published
|
|
3EWL
 
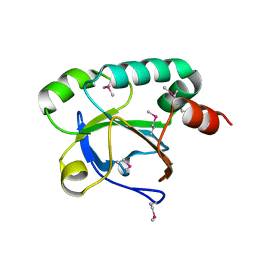 | |
3GE2
 
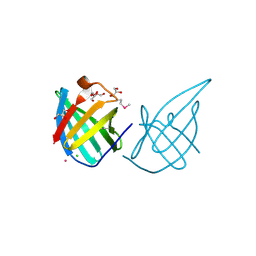 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
3FPI
 
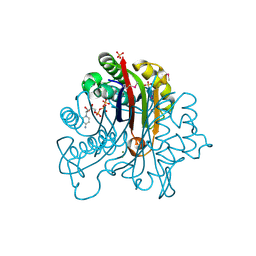 | | Crystal Structure of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase IspF complexed with Cytidine Triphosphate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of 2-C-Methyl-D-Erythritol 2,4-Cyclodiphosphate Synthase IspF complexed with Cytidine Triphosphate
To be Published
|
|
2EW2
 
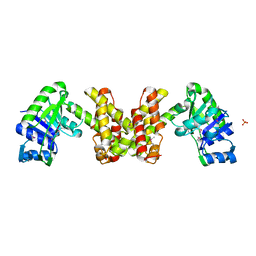 | | Crystal Structure of the Putative 2-Dehydropantoate 2-Reductase from Enterococcus faecalis | | Descriptor: | 2-dehydropantoate 2-reductase, putative, MAGNESIUM ION, ... | | Authors: | Kim, Y, Zhou, M, Moy, S, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative 2-Dehydropantoate 2-Reductase from Enterococcus faecalis
To be Published
|
|
3GVE
 
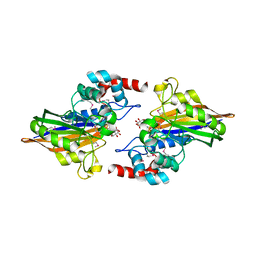 | | Crystal structure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis | | Descriptor: | CITRIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Kim, Y, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-28 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Calcineurin-like Phosphoesterase from Bacillus subtilis
To be Published
|
|
2FA8
 
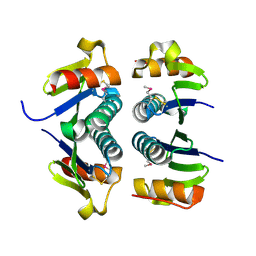 | | Crystal Structure of the Putative Selenoprotein W-related family Protein from Agrobacterium tumefaciens | | Descriptor: | hypothetical protein Atu0228 | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-07 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Putative Selenoprotein W-related family Protein from Agrobacterium tumefaciens
To be Published, 2006
|
|
2FIU
 
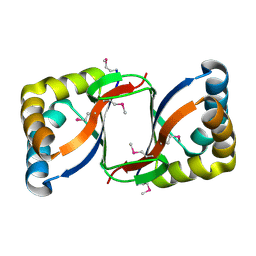 | | Crystal Structure of the Conserved Protein of Unknown Function ATU0297 from Agrobacterium tumefaciens | | Descriptor: | conserved hypothetical protein | | Authors: | Kim, Y, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Atu0297 from Agrobacterium tumefaciens
To be Published
|
|
2FHQ
 
 | |
2FHP
 
 | |
2FIW
 
 | | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris | | Descriptor: | ACETYL COENZYME *A, GCN5-related N-acetyltransferase:Aminotransferase, class-II, ... | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris
To be Published, 2006
|
|
8H8V
 
 | |
8H8U
 
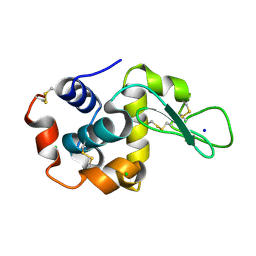 | |
8H8T
 
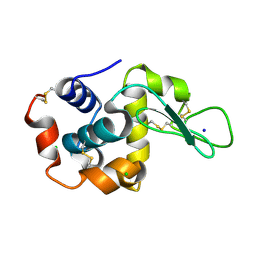 | |
8H8W
 
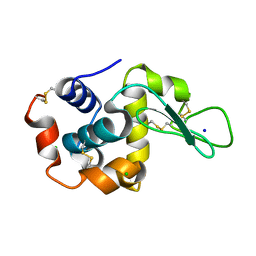 | |
3LNO
 
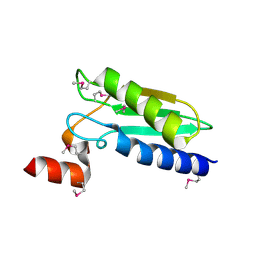 | | Crystal Structure of Domain of Unknown Function DUF59 from Bacillus anthracis | | Descriptor: | Putative uncharacterized protein | | Authors: | Kim, Y, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-02 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Domain of Unknown Function DUF59 Bacillus anthracis
To be Published
|
|
2I7G
 
 | | Crystal Structure of Monooxygenase from Agrobacterium tumefaciens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Monooxygenase, SULFATE ION | | Authors: | Kim, Y, Xu, X, Zheng, H, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of Monooxygenase from Agrobacterium tumefaciens
To be Published, 2006
|
|
2I7H
 
 | | Crystal Structure of the Nitroreductase-like Family Protein from Bacillus cereus | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase-like family protein, SULFATE ION | | Authors: | Kim, Y, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nitroreductase-like Family Protein from Bacillus cereus
To be Published
|
|
3LVY
 
 | | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Carboxymuconolactone decarboxylase family, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans
To be Published
|
|
2IGT
 
 | | Crystal Structure of the SAM Dependent Methyltransferase from Agrobacterium tumefaciens | | Descriptor: | ACETIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Gu, J, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-25 | | Release date: | 2006-10-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the SAM Dependent Methyltransferase from Agrobacterium tumefaciens
To be Published
|
|
3NAT
 
 | | Crystal Structure of Conserved Protein of Unknown Function EF_1977 from Enterococcus faecalis | | Descriptor: | CITRIC ACID, Uncharacterized protein, ZINC ION | | Authors: | Kim, Y, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.925 Å) | | Cite: | Crystal Structure of Conserved Protein of Unknown Function EF_1977 from Enterococcus faecalis
To be Published
|
|
2IGS
 
 | | Crystal Structure of the Protein of Unknown Function from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, GLYCEROL, Hypothetical protein, ... | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Egorova, O, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-25 | | Release date: | 2006-10-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Pseudomonas aeruginosa
To be Published
|
|
3NA2
 
 | | Crystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crsystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum
To be Published
|
|
3NKH
 
 | | Crystal Structure of Integrase from MRSA strain Staphylococcus aureus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-19 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal Structure of Integrase from MRSA strain Staphylococcus aureus
To be Published, 2010
|
|
2IQQ
 
 | | The Crystal Structure of Iron, Sulfur-Dependent L-serine dehydratase from Legionella pneumophila subsp. pneumophila | | Descriptor: | Iron, Sulfur-Dependent L-serine dehydratase, MAGNESIUM ION | | Authors: | Kim, Y, Hatzos, C, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-14 | | Release date: | 2006-11-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The Crystal Structure of Iron, Sulfur-Dependent L-serine dehydratase from Legionella pneumophila subsp. pneumophila
To be Published
|
|
