8TWT
 
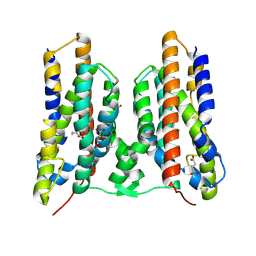 | |
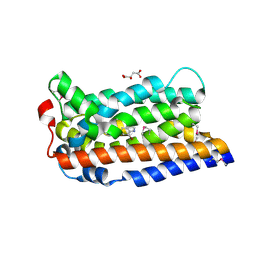
8TWN
 
 | |
8TWW
 
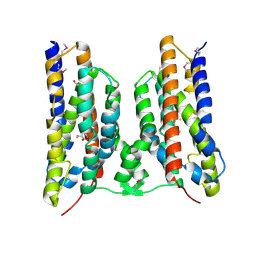 | |
4HHY
 
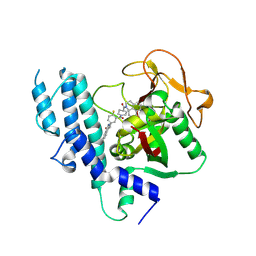 | | Crystal structure of PARP catalytic domain in complex with novel inhibitors | | Descriptor: | (9aR)-1-[(1-{2-fluoro-5-[(4-oxo-3,4-dihydrophthalazin-1-yl)methyl]benzoyl}piperidin-4-yl)carbonyl]-1,2,3,8,9,9a-hexahydro-7H-benzo[de][1,7]naphthyridin-7-one, DI(HYDROXYETHYL)ETHER, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-10-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3637 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of a Series of Benzo[de][1,7]naphthyridin-7(8H)-ones Bearing a Functionalized Longer Chain Appendage as Novel PARP1 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4HHZ
 
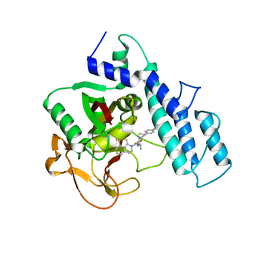 | | Crystal structure of PARP catalytic domain in complex with novel inhibitors | | Descriptor: | N-{(2S)-1-[4-(4-fluorophenyl)-3,6-dihydropyridin-1(2H)-yl]-1-oxopropan-2-yl}-2-[(9aR)-7-oxo-2,3,7,8,9,9a-hexahydro-1H-benzo[de][1,7]naphthyridin-1-yl]acetamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-10-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7199 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of a Series of Benzo[de][1,7]naphthyridin-7(8H)-ones Bearing a Functionalized Longer Chain Appendage as Novel PARP1 Inhibitors.
J.Med.Chem., 56, 2013
|
|
3PHS
 
 | |
4PY4
 
 | |
