3CUN
 
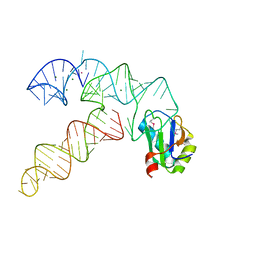 | | Aminoacyl-tRNA synthetase ribozyme | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Xiao, H, Murakami, H, Suga, H, Ferre-D'Amare, A.R. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Nature, 454, 2008
|
|
9LR9
 
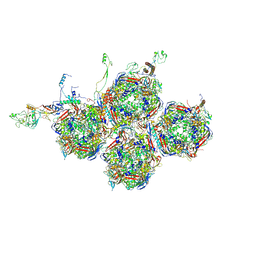 | | Local reconstruction of bovine adenovirus type 3 capsid | | Descriptor: | Hexon protein, PIX, PV, ... | | Authors: | Xiao, H, Liu, H.R. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Insights into Minor and Core Proteins of Bovine Adenovirus 3: Bridging Capsid and Genomic Core.
J.Mol.Biol., 437, 2025
|
|
9LTA
 
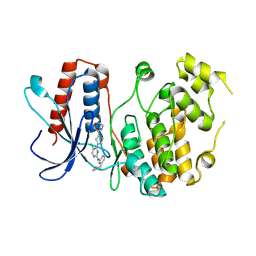 | | Crystal Structure of Compound SKLB-D18 with MAPK7 (ERK5) | | Descriptor: | 4-[5-chloranyl-2-[[3-[(dimethylamino)methyl]phenyl]amino]pyrimidin-4-yl]-~{N}-morpholin-4-yl-thiophene-2-carboxamide, Mitogen-activated protein kinase 7 | | Authors: | Xiao, H, Sun, Q. | | Deposit date: | 2025-02-05 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A first-in-class selective inhibitor of ERK1/2 and ERK5 overcomes drug resistance with a single-molecule strategy.
Signal Transduct Target Ther, 10, 2025
|
|
9LNR
 
 | | Crystal structure of SKLB-D18 with ERK2 | | Descriptor: | 4-[5-chloranyl-2-[[3-[(dimethylamino)methyl]phenyl]amino]pyrimidin-4-yl]-~{N}-morpholin-4-yl-thiophene-2-carboxamide, GLYCEROL, Mitogen-activated protein kinase 1 | | Authors: | Xiao, H, Sun, Q. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A first-in-class selective inhibitor of ERK1/2 and ERK5 overcomes drug resistance with a single-molecule strategy.
Signal Transduct Target Ther, 10, 2025
|
|
4ZJ1
 
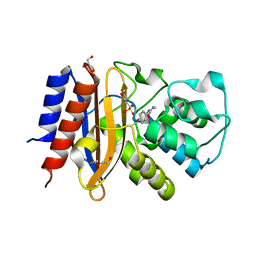 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli : V216AcrF mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase TEM, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZJ2
 
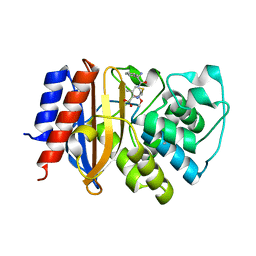 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli :E166N mutant | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, C.S, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZJ3
 
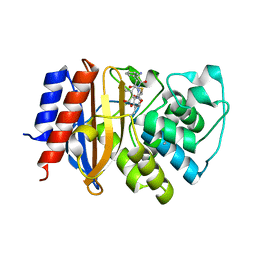 | | Crystal structure of cephalexin bound acyl-enzyme intermediate of Val216AcrF mutant TEM1 beta-lactamase from Escherichia coli: E166N and V216AcrF mutant. | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3EGZ
 
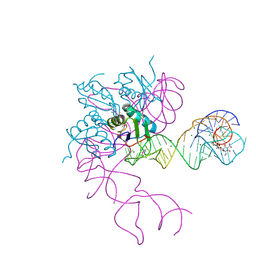 | | Crystal structure of an in vitro evolved tetracycline aptamer and artificial riboswitch | | Descriptor: | 7-CHLOROTETRACYCLINE, MAGNESIUM ION, Tetracycline aptamer and artificial riboswitch, ... | | Authors: | Xiao, H, Edwards, T.E, Ferre-D'Amare, A.R. | | Deposit date: | 2008-09-11 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch
Chem.Biol., 15, 2008
|
|
3CUL
 
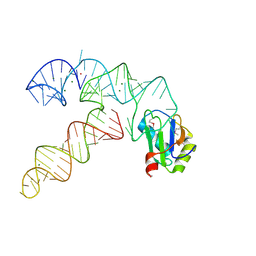 | | Aminoacyl-tRNA synthetase ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (92-MER), ... | | Authors: | Xiao, H, Murakami, H, Suga, H, Ferre-D'Amare, A.R. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Nature, 454, 2008
|
|
8I1V
 
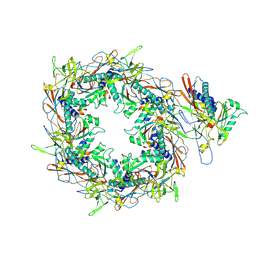 | | The asymmetric unit of P22 procapsid | | Descriptor: | Major capsid protein, Scaffolding protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
8I1T
 
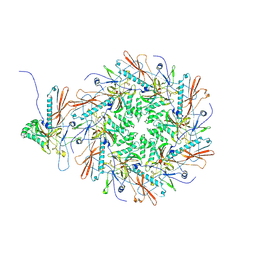 | | The asymmetric unit of P22 empty capsid | | Descriptor: | Major capsid protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
5Y2K
 
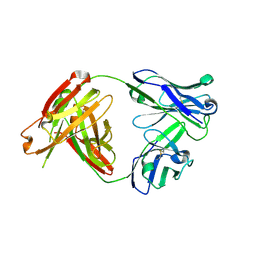 | | A human antibody AF4H1L1 | | Descriptor: | a group 2 HA binding antibody AF4H1K1 Fab heavy chain, a group 2 HA binding antibody AF4H1K1 Fab light chain | | Authors: | Xiao, H, Qi, J, Gao, F.G. | | Deposit date: | 2017-07-26 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An H3-clade neutralizing antibody screened from an H7N9 patient that binds group 2 influenza A hemagglutinins
To Be Published
|
|
5Y2M
 
 | |
5Y2L
 
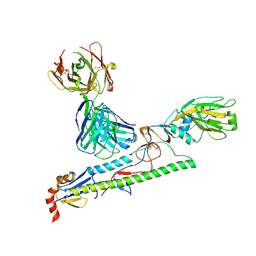 | |
8K36
 
 | |
8K35
 
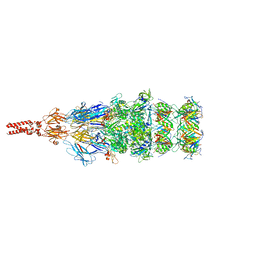 | | Structure of the bacteriophage lambda tail tip complex | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K37
 
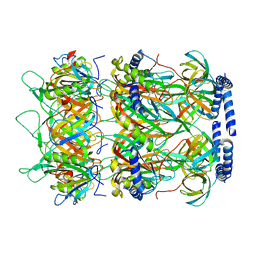 | | Structure of the bacteriophage lambda neck | | Descriptor: | Head-tail connector protein FII, Tail tube protein, Tail tube terminator protein | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K38
 
 | | The structure of bacteriophage lambda portal-adaptor | | Descriptor: | Head completion protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K39
 
 | | Structure of the bacteriophage lambda portal vertex | | Descriptor: | Major capsid protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
7E4K
 
 | |
9JG6
 
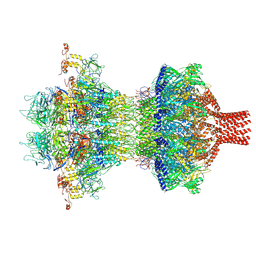 | | The tail-complex structure of phage P22 | | Descriptor: | Endorhamnosidase, P22 tail accessory factor, Phage stabilisation protein, ... | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-09-06 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYV
 
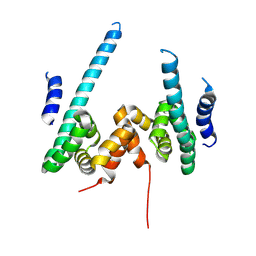 | | The scaffold dimer of phage P22 | | Descriptor: | Scaffolding protein | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-12-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYW
 
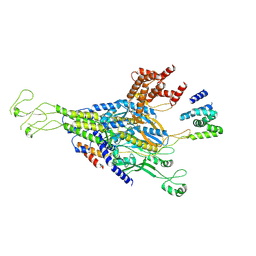 | | The scaffold C-loop of phage P22 | | Descriptor: | Portal protein, Scaffolding protein | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-12-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9JGA
 
 | | P22 procapsid portal | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-09-06 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYY
 
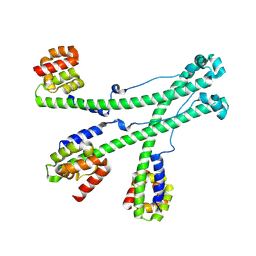 | | The scaffold trimer of phage P22 | | Descriptor: | Scaffolding protein | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-12-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
