1HF4
 
 | | STRUCTURAL EFFECTS OF MONOVALENT ANIONS ON POLYMORPHIC LYSOZYME CRYSTALS | | Descriptor: | LYSOZYME, NITRATE ION, SODIUM ION | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 2000-11-29 | | Release date: | 2001-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Effects of Monovalent Anions on Polymorphic Lysozyme Crystals
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7QRG
 
 | |
2V33
 
 | |
7QRE
 
 | | Structure of the hetero-tetramer complex between precursor membrane protein fragment (pr) and envelope protein (E) from tick-borne encephalitis virus | | Descriptor: | ACETATE ION, Envelope protein E, Genome polyprotein, ... | | Authors: | Vaney, M.C, Dellarole, M, Rey, F.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery.
Nat Commun, 13, 2022
|
|
7QRF
 
 | | Structure of the dimeric complex between precursor membrane ectodomain (prM) and envelope protein ectodomain (E) from tick-borne encephalitis virus | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Envelope protein E, ... | | Authors: | Vaney, M.C, Rouvinski, A, Rey, F.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery.
Nat Commun, 13, 2022
|
|
1B2K
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | IODIDE ION, PROTEIN (LYSOZYME) | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-26 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1B0D
 
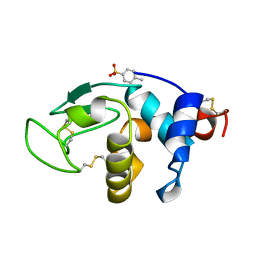 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | LYSOZYME, PARA-TOLUENE SULFONATE | | Authors: | Vaney, M.C, Broutin, I, Retailleau, P, Lafont, S, Hamiaux, C, Prange, T, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-07 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
194L
 
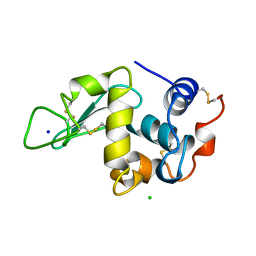 | | THE 1.40 A STRUCTURE OF SPACEHAB-01 HEN EGG WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Vaney, M.C, Maignan, S, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
193L
 
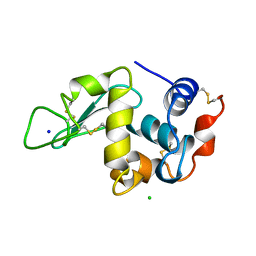 | | THE 1.33 A STRUCTURE OF TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Vaney, M.C, Maignan, S, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
5N0A
 
 | | Crystal structure of A259C covalently linked dengue 2 virus envelope glycoprotein dimer in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 HEAVY CHAIN, Envelope Glycoprotein E, ... | | Authors: | Vaney, M.C, Rouvinski, A, Guardado-Calvo, P, Sharma, A, Rey, F.A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope.
Nat Commun, 8, 2017
|
|
5LBS
 
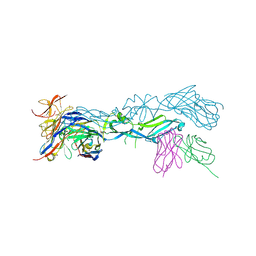 | | structural basis of Zika and Dengue virus potent antibody cross-neutralization | | Descriptor: | 1,2-ETHANEDIOL, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8, SULFATE ION, ... | | Authors: | Vaney, M.C, Rouvinski, A, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
6S8C
 
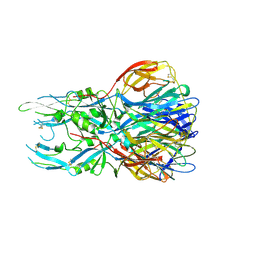 | |
4B3V
 
 | | Crystal structure of the Rubella virus glycoprotein E1 in its post-fusion form crystallized in presence of 20mM of Calcium Acetate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Vaney, M.C, DuBois, R.M, Tortorici, M.A, Rey, F.A. | | Deposit date: | 2012-07-26 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Functional and Evolutionary Insight from the Crystal Structure of Rubella Virus Protein E1.
Nature, 493, 2013
|
|
1LCN
 
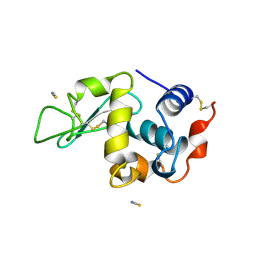 | | Monoclinic hen egg white lysozyme, thiocyanate complex | | Descriptor: | PROTEIN (LYSOZYME), THIOCYANATE ION | | Authors: | Hamiaux, C, Prange, T, Ducruix, A, Vaney, M.C. | | Deposit date: | 1998-10-27 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2ALA
 
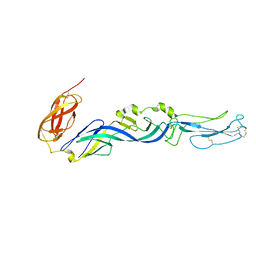 | | Crystal structure of the Semliki Forest Virus envelope protein E1 in its monomeric conformation. | | Descriptor: | Structural polyprotein (P130) | | Authors: | Roussel, A, Lescar, J, Vaney, M.C, Wengler, G, Wengler, G, Rey, F.A. | | Deposit date: | 2005-08-05 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus.
Structure, 14, 2006
|
|
1U3I
 
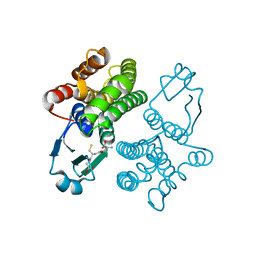 | | Crystal structure of glutathione S-tranferase from Schistosoma mansoni | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 28 kDa | | Authors: | Chomilier, J, Vaney, M.C, Labesse, G, Trottein, F, Capron, A, Mormon, J.-P. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Schistosoma mansoni glutathione S-transferase
To be Published
|
|
1RER
 
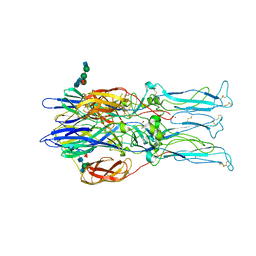 | | Crystal structure of the homotrimer of fusion glycoprotein E1 from Semliki Forest Virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gibbons, D.L, Vaney, M.C, Roussel, A, Vigouroux, A, Reilly, B, Kielian, M, Rey, F.A. | | Deposit date: | 2003-11-07 | | Release date: | 2004-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus.
Nature, 427, 2004
|
|
1UTG
 
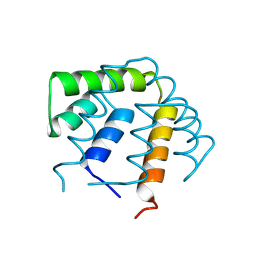 | | REFINEMENT OF THE C2221 CRYSTAL FORM OF OXIDIZED UTEROGLOBIN AT 1.34 ANGSTROMS RESOLUTION | | Descriptor: | UTEROGLOBIN | | Authors: | Morize, I, Surcouf, E, Vaney, M.C, Buehner, M, Mornon, J.P. | | Deposit date: | 1989-04-03 | | Release date: | 1989-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Refinement of the C222(1) crystal form of oxidized uteroglobin at 1.34 A resolution.
J.Mol.Biol., 194, 1987
|
|
6RX1
 
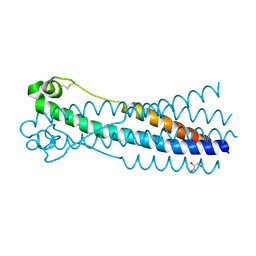 | | Crystal structure of human syncytin 1 in post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, Syncytin-1 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
6RX3
 
 | | Crystal structure of human syncytin 2 in post-fusion conformation | | Descriptor: | CHLORIDE ION, Syncytin-2 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
2XFC
 
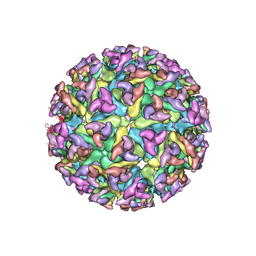 | | CHIKUNGUNYA E1 E2 ENVELOPE GLYCOPROTEINS FITTED IN SEMLIKI FOREST VIRUS cryo-EM MAP | | Descriptor: | E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Voss, J.E, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-Ray Crystallography
Nature, 468, 2010
|
|
2XT1
 
 | | Crystal structure of the HIV-1 capsid protein C-terminal domain (146- 231) in complex with a camelid VHH. | | Descriptor: | CAMELID VHH 9, GAG POLYPROTEIN, GLYCEROL | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-10-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2XXC
 
 | | Crystal structure of a camelid VHH raised against the HIV-1 capsid protein C-terminal domain. | | Descriptor: | CAMELID VHH 9 | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2XXM
 
 | | Crystal structure of the HIV-1 capsid protein C-terminal domain in complex with a camelid VHH and the CAI peptide. | | Descriptor: | ACETATE ION, CAMELID VHH 9, CAPSID PROTEIN P24, ... | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
2XV6
 
 | | Crystal structure of the HIV-1 capsid protein C-terminal domain (146- 220) in complex with a camelid VHH. | | Descriptor: | CAMELID VHH 9, CAPSID PROTEIN P24 | | Authors: | Igonet, S, Vaney, M.C, Bartonova, V, Helma, J, Rothbauer, U, Leonhardt, H, Stura, E, Krausslich, H.-G, Rey, F.A. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Targeting HIV-1 Virion Formation with Nanobodies -Implications for the Design of Assembly Inhibitors
To be Published
|
|
