6ZBX
 
 | | Structure of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6- alpha-manno-cyclophellitol carbasugar-stabilised trisaccharide inhibitor | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, (1~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, ... | | Authors: | Schroeder, S, Offen, W.A, Jin, Y, De Boer, C, Enoterpi, J, Marino, L, van der Marel, G.A, Codee, J.D.C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
7NL5
 
 | | Structure of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol trisaccharide inhibitor | | Descriptor: | (1R,2R,3R,4S,5R)-4-(hydroxymethyl)cyclohexane-1,2,3,5-tetrol, (1R,6S)-5beta-(Hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2beta,3beta,4alpha-triol, Alpha-1,6-mannanase, ... | | Authors: | Schroeder, S, Offen, W.A, Males, A, Jin, Y, De Boer, C, Enotarpi, J, Marino, L, van der Marel, G.A, Florea, B.I, Codee, J.D.C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
1NDN
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1992-01-15 | | Release date: | 1992-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
4INR
 
 | | Yeast 20S proteasome in complex with the vinyl sulfone LU102 | | Descriptor: | N3Phe-Leu-Leu-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | Authors: | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | Deposit date: | 2013-01-06 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
4INT
 
 | | Yeast 20S proteasome in complex with the vinyl sulfone LU122 | | Descriptor: | HMB-Val-Ser-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | Authors: | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | Deposit date: | 2013-01-06 | | Release date: | 2013-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
4INU
 
 | | Yeast 20S proteasome in complex with the vinyl sulfone LU112 | | Descriptor: | N3Phe-Phe(4-NH2CH2)-Leu-Phe(4-NH2CH2)-methyl vinyl sulfone, bound form, Proteasome component C1, ... | | Authors: | Geurink, P.P, van der Linden, W.A, Mirabella, A.C, Gallastegui, N, de Bruin, G, Blom, A.E.M, Voges, M.J, Mock, E.D, Florea, B.I, van der Marel, G.A, Driessen, C, van der Stelt, M, Groll, M, Overkleeft, H.S, Kisselev, A.F. | | Deposit date: | 2013-01-06 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites.
J.Med.Chem., 56, 2013
|
|
1UZ8
 
 | | anti-Lewis X Fab fragment in complex with Lewis X | | Descriptor: | IGG FAB (IGG3, KAPPA) HEAVY CHAIN 291-2G3-A, KAPPA) LIGHT CHAIN 291-2G3-A, ... | | Authors: | Van Roon, A.M.M, Pannu, N.S, De Vrind, J.P.M, Hokke, C.H, Deelder, A.M, Van Der marel, G.A, Van Boom, J.H, Abrahams, J.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an Anti-Lewis X Fab Fragment in Complex with its Lewis X Antigen
Structure, 12, 2004
|
|
1UZ6
 
 | | anti-Lewis X Fab fragment uncomplexed | | Descriptor: | IGG FAB (IGG3, KAPPA) HEAVY CHAIN 291-2G3-A, KAPPA) LIGHT CHAIN 291-2G3-A, ... | | Authors: | Van Roon, A.M.M, Pannu, N.S, De Vrind, J.P.M, Hokke, C.H, Deelder, A.M, Van Der marel, G.A, Van Boom, J.H, Abrahams, J.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an Anti-Lewis X Fab Fragment in Complex with its Lewis X Antigen
Structure, 12, 2004
|
|
4DNB
 
 | | METHYLATION OF THE ECORI RECOGNITION SITE DOES NOT ALTER DNA CONFORMATION. THE CRYSTAL STRUCTURE OF D(CGCGAM6ATTCGCG) AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*(6MA)P*TP*TP*CP*GP*CP*G)-3') | | Authors: | Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Methylation of the EcoRI recognition site does not alter DNA conformation: the crystal structure of d(CGCGAm6ATTCGCG) at 2.0-A resolution.
J.Biol.Chem., 263, 1988
|
|
1VTY
 
 | | Crystal structure of a Z-DNA fragment containing thymine/2-aminoadenine base pairs | | Descriptor: | AMINO GROUP, DNA (5'-D(*CP*(NH2)AP*CP*GP*TP*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Wang, A.H.-J, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a Z-DNA fragment containing thymine/2-aminoadenine base pairs.
J. Biomol. Struct. Dyn., 4, 1986
|
|
1O55
 
 | | MOLECULAR STRUCTURE OF TWO CRYSTAL FORMS OF CYCLIC TRIADENYLIC ACID AT 1 ANGSTROM RESOLUTION | | Descriptor: | COBALT (II) ION, DNA (5'-CD(*AP*AP*AP)-3') | | Authors: | Gao, Y.G, Robinson, H, Guan, Y, Liaw, Y.C, van Boom, J.H, van der Marel, G.A, Wang, A.H. | | Deposit date: | 2003-08-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Molecular structure of two crystal forms of cyclic triadenylic acid at 1A resolution.
J.Biomol.Struct.Dyn., 16, 1998
|
|
1O56
 
 | | MOLECULAR STRUCTURE OF TWO CRYSTAL FORMS OF CYCLIC TRIADENYLIC ACID AT 1 ANGSTROM RESOLUTION | | Descriptor: | DNA (5'-CD(*AP*AP*AP*)-3') | | Authors: | Gao, Y.G, Robinson, H, Guan, Y, Liaw, Y.C, van Boom, J.H, van der Marel, G.A, Wang, A.H. | | Deposit date: | 2003-08-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Molecular structure of two crystal forms of cyclic triadenylic acid at 1A resolution.
J.Biomol.Struct.Dyn., 16, 1998
|
|
308D
 
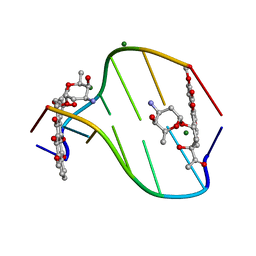 | | BINDING OF DAUNOMYCIN TO B-D-GLUCOSYLATED DNA FOUND IN PROTOZOA TRYPANOSOMA BRUCEI STUDIED BY X-RAY CRYSTALLOGRAPH | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Gao, Y.-G, Robinson, H, Wijsman, E.R, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1997-01-15 | | Release date: | 1997-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of Daunomycin to beta-D-Glucosylated DNA Found in Protozoa Trypanosoma brucei Studied by X-Ray Crystallography
J.Am.Chem.Soc., 119, 1997
|
|
1D13
 
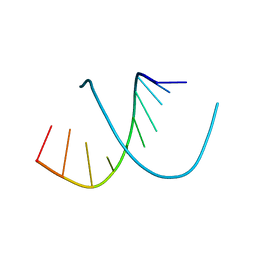 | | MOLECULAR STRUCTURE OF AN A-DNA DECAMER D(ACCGGCCGGT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Frederick, C.A, Quigley, G.J, Teng, M.-K, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of an A-DNA decamer d(ACCGGCCGGT).
Eur.J.Biochem., 181, 1989
|
|
1VTG
 
 | | THE MOLECULAR STRUCTURE OF A DNA-TRIOSTIN A COMPLEX | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), triostin A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular structure of a DNA-triostin A complex.
Science, 225, 1984
|
|
1VTE
 
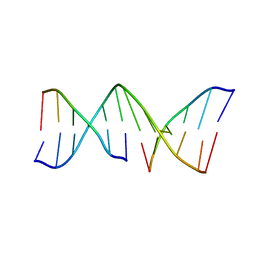 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL A4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1990-05-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc. Natl. Acad. Sci. U.S.A., 87, 1990
|
|
264D
 
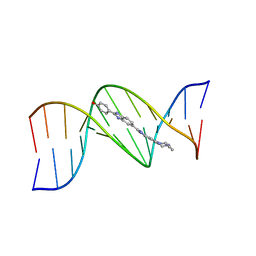 | | THREE-DIMENSIONAL CRYSTAL STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR-GROOVE-BINDING DRUG HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Vega, M.C, Garcia-Saez, I, Aymami, J, Eritja, R, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Coll, M. | | Deposit date: | 1994-09-22 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Three-dimensional crystal structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor-groove-binding drug Hoechst 33258.
Eur.J.Biochem., 222, 1994
|
|
1D14
 
 | | STRUCTURE OF 11-DEOXYDAUNOMYCIN BOUND TO DNA CONTAINING A PHOSPHOROTHIOATE | | Descriptor: | 6-DEOXYDAUNOMYCIN, DNA (5'-D(*CP*GP*TP*(AS)P*CP*G)-3') | | Authors: | Williams, L.D, Egli, M, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Quigley, G.J, Wang, A.H.-J, Rich, A, Frederick, C.A. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 11-deoxydaunomycin bound to DNA containing a phosphorothioate.
J.Mol.Biol., 215, 1990
|
|
1D12
 
 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), DOXORUBICIN, SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
1DNE
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 2 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-09-14 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment.
Biochemistry, 28, 1989
|
|
1D10
 
 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
1B4Y
 
 | | STRUCTURE AND MECHANISM OF FORMATION OF THE H-Y5 ISOMER OF AN INTRAMOLECULAR DNA TRIPLE HELIX. | | Descriptor: | DNA (H-Y5 TRIPLE HELIX) | | Authors: | Van Dongen, M.J.P, Doreleijers, J.F, Van Der Marel, G.A, Van Boom, J.H, Hilbers, C.W, Wijmenga, S.S. | | Deposit date: | 1998-12-30 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of formation of the H-y5 isomer of an intramolecular DNA triple helix.
Nat.Struct.Biol., 6, 1999
|
|
1AC7
 
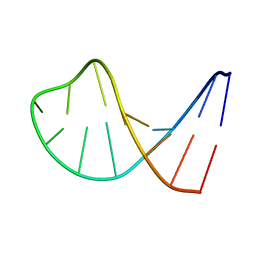 | | STRUCTURAL FEATURES OF THE DNA HAIRPIN D(ATCCTAGTTATAGGAT): THE FORMATION OF A G-A BASE PAIR IN THE LOOP, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*TP*AP*GP*TP*TP*AP*TP*AP*GP*GP*AP*T)-3') | | Authors: | Van Dongen, M.J.P, Mooren, M.M.W, Willems, E.F.A, Van Der Marel, G.A, Van Boom, J.H, Wijmenga, S.S, Hilbers, C.W. | | Deposit date: | 1997-02-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural features of the DNA hairpin d(ATCCTA-GTTA-TAGGAT): formation of a G-A base pair in the loop.
Nucleic Acids Res., 25, 1997
|
|
121D
 
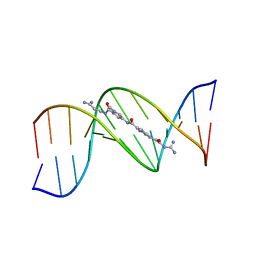 | | MOLECULAR STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR GROOVE BINDING DRUG NETROPSIN | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), NETROPSIN | | Authors: | Tabernero, L, Verdaguer, N, Coll, M, Fita, I, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Aymami, J. | | Deposit date: | 1993-04-14 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor groove binding drug netropsin.
Biochemistry, 32, 1993
|
|
128D
 
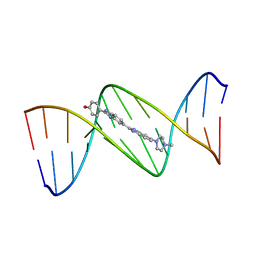 | | MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG) COMPLEXED WITH HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1993-06-30 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
