1CBG
 
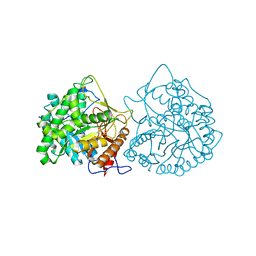 | | THE CRYSTAL STRUCTURE OF A CYANOGENIC BETA-GLUCOSIDASE FROM WHITE CLOVER (TRIFOLIUM REPENS L.), A FAMILY 1 GLYCOSYL-HYDROLASE | | Descriptor: | CYANOGENIC BETA-GLUCOSIDASE | | Authors: | Barrett, T.E, Suresh, C.G, Tolley, S.P, Hughes, M.A. | | Deposit date: | 1995-07-31 | | Release date: | 1995-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a cyanogenic beta-glucosidase from white clover, a family 1 glycosyl hydrolase.
Structure, 3, 1995
|
|
3ENG
 
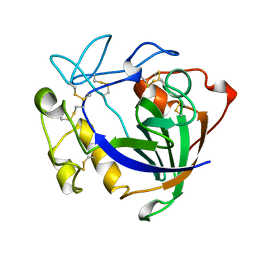 | | STRUCTURE OF ENDOGLUCANASE V CELLOBIOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOBIOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1AI5
 
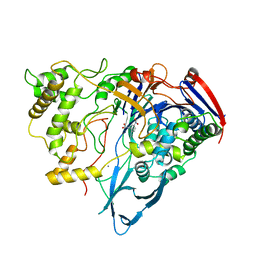 | | PENICILLIN ACYLASE COMPLEXED WITH M-NITROPHENYLACETIC ACID | | Descriptor: | 2-(3-NITROPHENYL)ACETIC ACID, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
4ENG
 
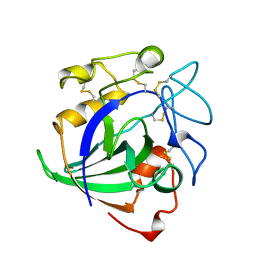 | | STRUCTURE OF ENDOGLUCANASE V CELLOHEXAOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOHEXAOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1TGL
 
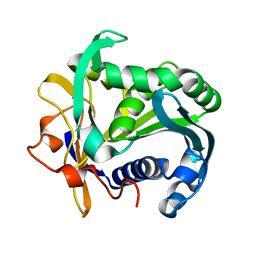 | | A SERINE PROTEASE TRIAD FORMS THE CATALYTIC CENTRE OF A TRIACYLGLYCEROL LIPASE | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1990-02-05 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A serine protease triad forms the catalytic centre of a triacylglycerol lipase.
Nature, 343, 1990
|
|
2ENG
 
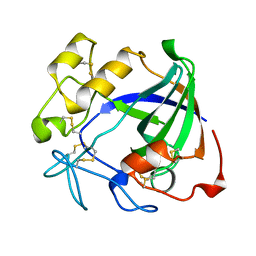 | | ENDOGLUCANASE V | | Descriptor: | ENDOGLUCANASE V | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1995-06-29 | | Release date: | 1996-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of oligosaccharide-bound forms of the endoglucanase V from Humicola insolens at 1.9 A resolution.
Biochemistry, 34, 1995
|
|
3TGL
 
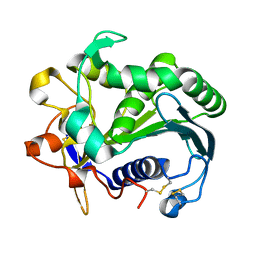 | | STRUCTURE AND MOLECULAR MODEL REFINEMENT OF RHIZOMUCOR MIEHEI TRIACYLGLYCERIDE LIPASE: A CASE STUDY OF THE USE OF SIMULATED ANNEALING IN PARTIAL MODEL REFINEMENT | | Descriptor: | TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURE AND MOLECULAR-MODEL REFINEMENT OF RHIZOMUCOR-MIEHEI TRIACYLGLYCERIDE LIPASE - A CASE-STUDY OF THE USE OF SIMULATED ANNEALING IN PARTIAL MODEL REFINEMENT.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1PNK
 
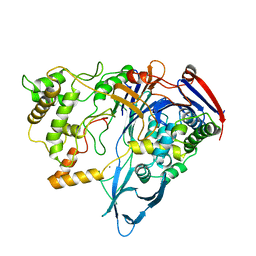 | |
1PNM
 
 | |
1PNL
 
 | |
1GM9
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-12 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GM7
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GM8
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GKF
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-13 | | Release date: | 2002-01-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1AI7
 
 | | PENICILLIN ACYLASE COMPLEXED WITH PHENOL | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, PHENOL | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AJN
 
 | | PENICILLIN ACYLASE COMPLEXED WITH P-NITROPHENYLACETIC ACID | | Descriptor: | 2-(4-NITROPHENYL)ACETIC ACID, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AJP
 
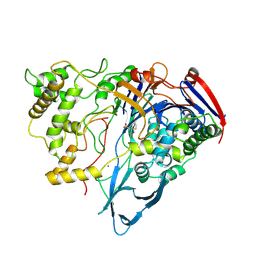 | |
1AJQ
 
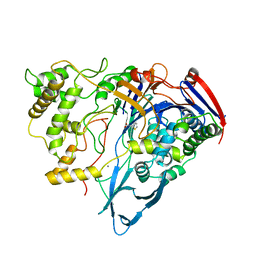 | | PENICILLIN ACYLASE COMPLEXED WITH THIOPHENEACETIC ACID | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, THIOPHENEACETIC ACID | | Authors: | Done, S.H. | | Deposit date: | 1997-05-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AI6
 
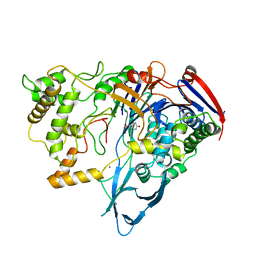 | | PENICILLIN ACYLASE WITH P-HYDROXYPHENYLACETIC ACID | | Descriptor: | 4-HYDROXYPHENYLACETATE, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AI4
 
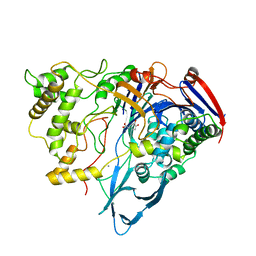 | |
1PID
 
 | | BOVINE DESPENTAPEPTIDE INSULIN | | Descriptor: | DESPENTAPEPTIDE INSULIN | | Authors: | Holden, P.H, Papiz, M, Dodson, G.G. | | Deposit date: | 1995-11-22 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A model of insulin fibrils derived from the x-ray crystal structure of a monomeric insulin (despentapeptide insulin).
Proteins, 27, 1997
|
|
1H2G
 
 | | Altered substrate specificity mutant of penicillin acylase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Morillas, M, Brannigan, J.A, Ladurner, A.G, Forney, L.J, Virden, R. | | Deposit date: | 2002-08-08 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of Penicillin Acylase Residue B71 Extend Substrate Specificity by Decreasing Steric Constraints for Substrate Binding
Biochem.J., 371, 2003
|
|
1GK9
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
