1RWD
 
 | |
2JN7
 
 | | Northeast Structural Genomics Consortium Target ER411 | | Descriptor: | protein yfjZ | | Authors: | Tian, F, Zhao, L, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-28 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of E.Coli hypothetical protein YFJZ
To be Published
|
|
8E22
 
 | | VPS37A_21-148 | | Descriptor: | Vacuolar protein sorting-associated protein 37A | | Authors: | Tian, F, Ye, Y.S. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Identification of membrane curvature sensing motifs essential for VPS37A phagophore recruitment and autophagosome closure.
Commun Biol, 7, 2024
|
|
2MVJ
 
 | |
2MVH
 
 | |
8FKM
 
 | |
1FH1
 
 | |
2JS1
 
 | | Solution NMR structure of the homodimer protein YVFG from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR478 | | Descriptor: | Uncharacterized protein yvfG | | Authors: | Macnaughtan, M.A, Weldeghiorghis, T, Wang, X, Bansal, S, Tian, F, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bacillus subtilis Protein YvfG.
To be Published
|
|
2K5C
 
 | | NMR Structure for PF0385 | | Descriptor: | ZINC ION, uncharacterized protein PF0385 | | Authors: | Prestegard, J.H, Tian, F, Baucom, C, Adams, M.W.W, Montelione, G.T, Sugar, F.J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of PF0385
To be Published
|
|
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | Descriptor: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
7ESF
 
 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
2G2R
 
 | |
8JAH
 
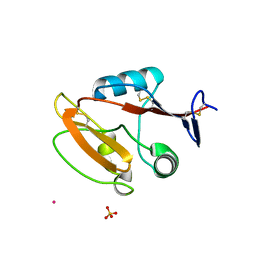 | | Crystal structure of human CLEC12A C-type lectin domain | | Descriptor: | C-type lectin domain family 12 member A, POTASSIUM ION, SULFATE ION | | Authors: | Lei, Q, Tang, H, Dong, X. | | Deposit date: | 2023-05-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Mechanistic insights into the C-type lectin receptor CLEC12A-mediated immune recognition of monosodium urate crystal.
J.Biol.Chem., 300, 2024
|
|
8I5M
 
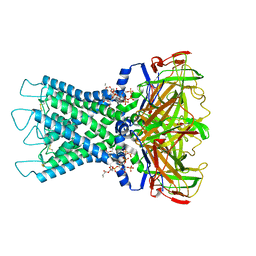 | | Rat Kir4.1 in complex with PIP2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
8I5N
 
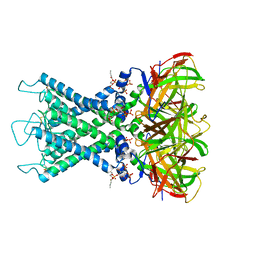 | | Rat Kir4.1 in complex with PIP2 and Lys05 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
8BBV
 
 | | Coproporphyrin III - LmCpfC complex soaked 2min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Coproporphyrin III ferrochelatase, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
8OFL
 
 | | Coproporphyrin III - LmCpfC complex soaked 4min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Coproporphyrin III ferrochelatase, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
8OMM
 
 | | Coproporphyrin III - LmCpfC complex soaked 3min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, Coproporphyrin III ferrochelatase, FE (II) ION, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
7OHD
 
 | | CRYSTAL STRUCTURE OF FERRIC MURINE NEUROGLOBIN CDLESS MUTANT | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Cerutti, G, Gugole, E, Vallone, B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Role of Murine Neuroglobin CDloop-D-Helix Unit in CO Ligand Binding and Structural Dynamics.
Acs Chem.Biol., 17, 2022
|
|
8AW7
 
 | | Structure of coproporphyrin III-LmCpfC R45L | | Descriptor: | Coproporphyrin III ferrochelatase, GLYCEROL, coproporphyrin III | | Authors: | Gabler, T, Hofbauer, S, Pfanzagl, V. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Active site architecture of coproporphyrin ferrochelatase with its physiological substrate coproporphyrin III: Propionate interactions and porphyrin core deformation.
Protein Sci., 32, 2023
|
|
8AT8
 
 | | Structure of coproporphyrin III-LmCpfC | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Active site architecture of coproporphyrin ferrochelatase with its physiological substrate coproporphyrin III: Propionate interactions and porphyrin core deformation.
Protein Sci., 32, 2023
|
|
6I6C
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 2 | | Descriptor: | (1~{R},2~{S},4~{S})-~{N}-(3-chloranyl-4-cyano-phenyl)sulfonylbicyclo[2.2.1]heptane-2-carboxamide, 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I6T
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1-oxidanyl-naphthalene-2-carboxylic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I79
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 6-[(4-~{tert}-butyl-1,3-thiazol-2-yl)methyl]-4,6-diazaspiro[2.4]heptane-5,7-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I6P
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 3 | | Descriptor: | 1,2-ETHANEDIOL, 6-azaspiro[3.4]octan-6-yl-[2,4-bis(chloranyl)-6-oxidanyl-phenyl]methanone, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
