2V6F
 
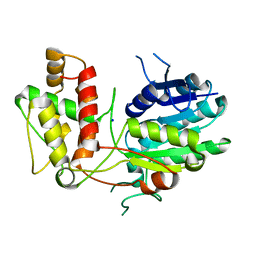 | | Structure of Progesterone 5beta-Reductase from Digitalis Lanata | | Descriptor: | PROGESTERONE 5-BETA-REDUCTASE, SODIUM ION | | Authors: | Thorn, A, Egerer-Sieber, C, Jaeger, C.M, Herl, V, Mueller-Uri, F, Kreis, W, Muller, Y.A. | | Deposit date: | 2007-07-18 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Progesterone 5{Beta}-Reductase from Digitalis Lanata Defines a Novel Class of Short Chain Dehydrogenases/Reductases.
J.Biol.Chem., 283, 2008
|
|
2V6G
 
 | | Structure of Progesterone 5beta-Reductase from Digitalis Lanata in complex with NADP | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE 5-BETA-REDUCTASE, ... | | Authors: | Thorn, A, Egerer-Sieber, C, Jaeger, C, Herl, V, Mueller-Uri, F, Kreis, W, Muller, Y.A. | | Deposit date: | 2007-07-18 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of Progesterone 5{Beta}-Reductase from Digitalis Lanata Defines a Novel Class of Short Chain Dehydrogenases/Reductases.
J.Biol.Chem., 283, 2008
|
|
3SZS
 
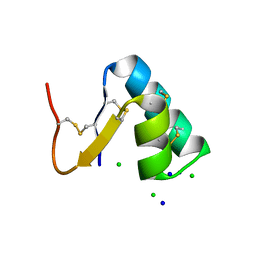 | |
3T0O
 
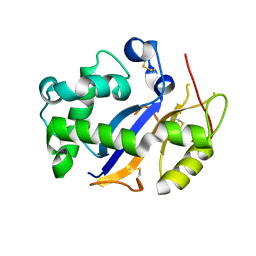 | | Crystal Structure Analysis of Human RNase T2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribonuclease T2 | | Authors: | Thorn, A, Kraetzner, R, Steinfeld, R, Sheldrick, G. | | Deposit date: | 2011-07-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and activity of the only human RNase T2.
Nucleic Acids Res., 40, 2012
|
|
8PFK
 
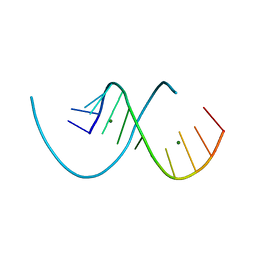 | | RNA structure with 1-methylpseudouridine, C2 space group | | Descriptor: | MAGNESIUM ION, RNA (12-mer) | | Authors: | Spingler, B, McAuley, K, Nievergelt, P, Thorn, A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.321 Å) | | Cite: | RNA oligomers at atomic resolution containing 1-methylpseudouridine, an essential building block of mRNA vaccines.
Chemmedchem, 19, 2024
|
|
8PFQ
 
 | | RNA structure with 1-methylpseudouridine, P1 space group | | Descriptor: | Chains: A,B, MAGNESIUM ION | | Authors: | Spingler, B, McAuley, K, Nievergelt, P, Thorn, A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | RNA oligomers at atomic resolution containing 1-methylpseudouridine, an essential building block of mRNA vaccines.
Chemmedchem, 19, 2024
|
|
4TS2
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, magnesium ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4TS0
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, barium ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, BARIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1BGS
 
 | | RECOGNITION BETWEEN A BACTERIAL RIBONUCLEASE, BARNASE, AND ITS NATURAL INHIBITOR, BARSTAR | | Descriptor: | BARNASE, BARSTAR | | Authors: | Guillet, V, Lapthorn, A, Mauguen, Y. | | Deposit date: | 1993-11-02 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Structure, 1, 1993
|
|
5IFM
 
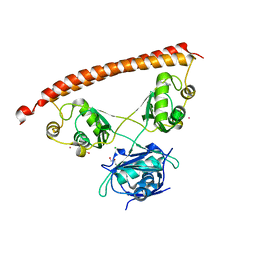 | | Human NONO (p54nrb) Homodimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-POU domain-containing octamer-binding protein, ... | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of human NONO (p54(nrb)): overcoming pathological problems with purification, data collection and noncrystallographic symmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4AUB
 
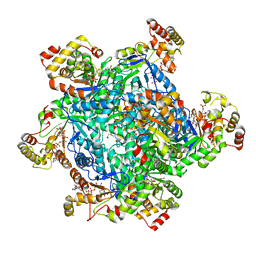 | |
4AST
 
 | |
2WON
 
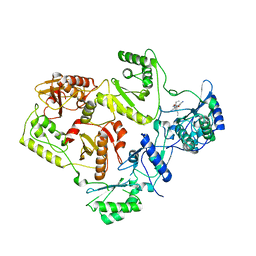 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (wild-type). | | Descriptor: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | Deposit date: | 2009-07-27 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lersivirine: A Non-Nucleoside Reverse Transcriptase Inhibitor with Activity Against Drug- Resistant Human Immunodeficiency Virus-1.
Antimicrob.Agents Chemother., 54, 2010
|
|
2WOM
 
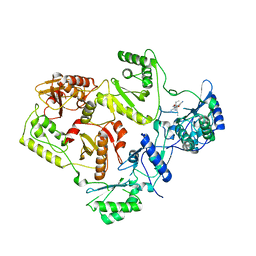 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (K103N). | | Descriptor: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | Deposit date: | 2009-07-27 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Lersivirine, a nonnucleoside reverse transcriptase inhibitor with activity against drug-resistant human immunodeficiency virus type 1.
Antimicrob. Agents Chemother., 54, 2010
|
|
1B2S
 
 | |
1BRS
 
 | |
1B3S
 
 | |
1B2U
 
 | |
1B27
 
 | |
