1VRV
 
 | |
2FEW
 
 | |
6ILU
 
 | | Endolysin LysPBC5 CBD | | Descriptor: | 1,2-ETHANEDIOL, Lysin, SULFATE ION | | Authors: | Suh, J.Y, Ryu, K.S, Ryu, S, Lee, K.O, Kong, M.S, Bae, J.W, Kim, I.T. | | Deposit date: | 2018-10-19 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural Basis for Cell-Wall Recognition by Bacteriophage PBC5 Endolysin.
Structure, 27, 2019
|
|
6JHW
 
 | | Structure of anti-CRISPR AcrIIC3 and NmeCas9 HNH | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9 | | Authors: | Suh, J.Y, Lee, B.J, Lee, S.J, Kim, Y. | | Deposit date: | 2019-02-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Anti-CRISPR AcrIIC3 discriminates between Cas9 orthologs via targeting the variable surface of the HNH nuclease domain.
Febs J., 286, 2019
|
|
6JHV
 
 | | Structure of anti-CRISPR AcrIIC3 | | Descriptor: | AcrIIC3 | | Authors: | Suh, J.Y, Lee, B.J, Lee, S.J, Kim, Y. | | Deposit date: | 2019-02-19 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Anti-CRISPR AcrIIC3 discriminates between Cas9 orthologs via targeting the variable surface of the HNH nuclease domain.
Febs J., 286, 2019
|
|
8JB9
 
 | |
6L5K
 
 | | ARF5 Aux/IAA17 Complex | | Descriptor: | Auxin response factor 5, Auxin-responsive protein IAA17 | | Authors: | Ryu, K.S, Suh, J.Y, Cha, S.Y, Kim, Y.I, Park, C.K. | | Deposit date: | 2019-10-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Determinants of PB1 Domain Interactions in Auxin Response Factor ARF5 and Repressor IAA17.
J.Mol.Biol., 432, 2020
|
|
6KZ7
 
 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | Descriptor: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | Deposit date: | 2019-09-23 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
6LZP
 
 | |
6LKF
 
 | |
6M3N
 
 | | Solution structure of anti-CRISPR AcrIF7 | | Descriptor: | anti-CRIPSR AcrIF7 | | Authors: | Kim, I, An, S.Y, Koo, J, Bae, E, Suh, J.Y. | | Deposit date: | 2020-03-04 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into the CRISPR inhibition of AcrIF7.
Nucleic Acids Res., 48, 2020
|
|
7VZM
 
 | | Anti-CRISPR AcrIE4-F7 | | Descriptor: | AcrIE4-F7 | | Authors: | Hong, S.H, Lee, G, Bae, E, Suh, J.Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of AcrIE4-F7 reveals a common strategy for dual CRISPR inhibition by targeting PAM recognition sites.
Nucleic Acids Res., 50, 2022
|
|
7XX9
 
 | |
7XX8
 
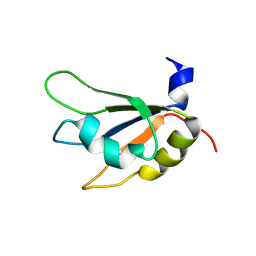 | |
5Y69
 
 | | Crystal structure of the L52M mutant of AcrIIA1 | | Descriptor: | chain A | | Authors: | Ka, D, An, S.Y, Suh, J.Y, Bae, E. | | Deposit date: | 2017-08-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an anti-CRISPR protein, AcrIIA1.
Nucleic Acids Res., 46, 2018
|
|
5Y6A
 
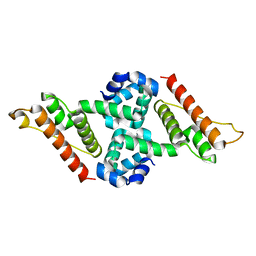 | | Crystal structure of the anti-CRISPR protein, AcrIIA1 | | Descriptor: | chain A and B | | Authors: | Ka, D, An, S.Y, Suh, J.Y, Bae, E. | | Deposit date: | 2017-08-11 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an anti-CRISPR protein, AcrIIA1
Nucleic Acids Res., 46, 2018
|
|
5XN4
 
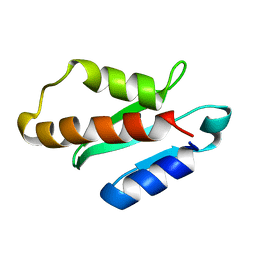 | | Anti-CRISPR protein AcrIIA4 | | Descriptor: | Anti-CRISPR AcrIIA4 | | Authors: | Suh, J.-Y, Kim, I. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor.
Sci Rep, 8, 2018
|
|
2XDF
 
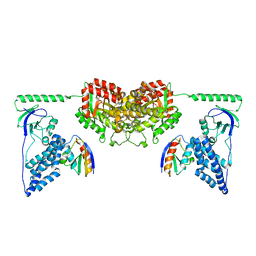 | | Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | Authors: | Schwieters, C.D, Suh, J.-Y, Grishaev, A, Guirlando, R, Takayama, Y, Clore, G.M. | | Deposit date: | 2010-04-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of the 128 kDa Enzyme I Dimer from Escherichia Coli and its 146 kDa Complex with Hpr Using Residual Dipolar Couplings and Small- and Wide-Angle X-Ray Scattering.
J.Am.Chem.Soc., 132, 2010
|
|
2KX9
 
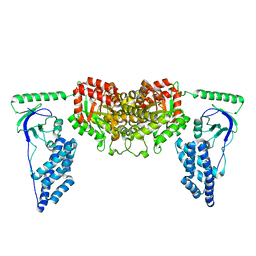 | | Solution Structure of the Enzyme I dimer Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Schwieters, C.D, Suh, J, Grishaev, A, Takayama, Y, Guirlando, R, Clore, G. | | Deposit date: | 2010-04-29 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering.
J.Am.Chem.Soc., 132, 2010
|
|
2MUK
 
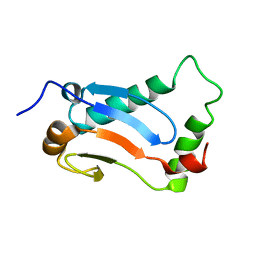 | |
2MNU
 
 | |
5H1P
 
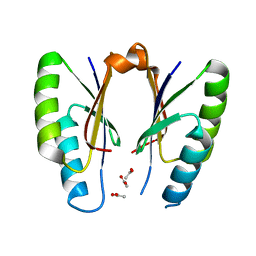 | | CRISPR-associated protein | | Descriptor: | ACETATE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Ka, D, Jeong, U, Bae, E. | | Deposit date: | 2016-10-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity
Struct Dyn, 4, 2017
|
|
5H1O
 
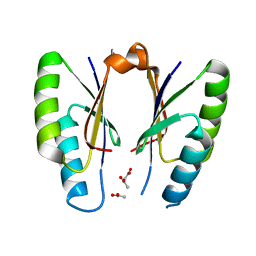 | | CRISPR-associated protein | | Descriptor: | ACETATE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Ka, D, Jeong, U, Bae, E. | | Deposit date: | 2016-10-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity
Struct Dyn, 4, 2017
|
|
8HEK
 
 | |
8IUD
 
 | |
