2V6V
 
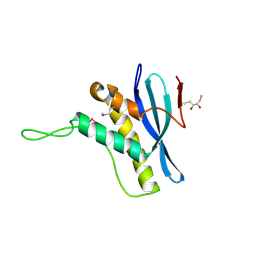 | | The structure of the Bem1p PX domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BUD EMERGENCE PROTEIN 1 | | Authors: | Stahelin, R.V, Karathanassis, D, Murray, D, Williams, R.L, Cho, W. | | Deposit date: | 2007-07-21 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Membrane Binding Analysis of the Phox Homology Domain of Bem1P: Basis of Phosphatidylinositol 4-Phosphate Specificity.
J.Biol.Chem., 282, 2007
|
|
2IWL
 
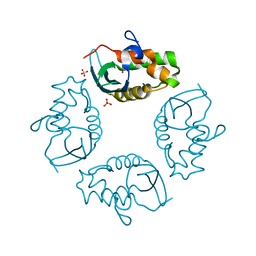 | |
5B0V
 
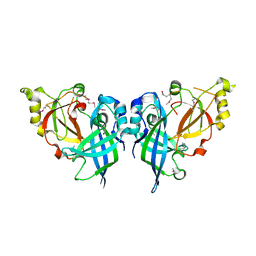 | | Crystal Structure of Marburg virus VP40 Dimer | | Descriptor: | ETHANOL, Matrix protein VP40 | | Authors: | Oda, S, Bornholdt, Z.A, Abelson, D.M, Saphire, E.O. | | Deposit date: | 2015-11-05 | | Release date: | 2016-01-20 | | Last modified: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Marburg Virus VP40 Reveals a Broad, Basic Patch for Matrix Assembly and a Requirement of the N-Terminal Domain for Immunosuppression
J.Virol., 90, 2015
|
|
3RCP
 
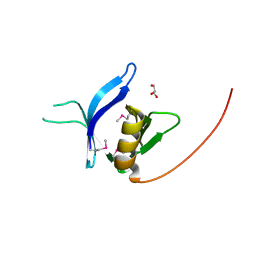 | | Crystal structure of the FAPP1 pleckstrin homology domain | | Descriptor: | GLYCEROL, Pleckstrin homology domain-containing family A member 3 | | Authors: | Roy, S, He, J, Kutateladze, T.G. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of Phosphatidylinositol 4-Phosphate and ARF1 GTPase Recognition by the FAPP1 Pleckstrin Homology (PH) Domain.
J.Biol.Chem., 286, 2011
|
|
6VSK
 
 | | Crystal structure of the P-Rex1 DEP1 domain | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Tesmer, J, Ravala, S.K. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The first DEP domain of the RhoGEF P-Rex1 autoinhibits activity and contributes to membrane binding.
J.Biol.Chem., 295, 2020
|
|
1O7K
 
 | | human p47 PX domain complex with sulphates | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 1, SULFATE ION | | Authors: | Karathanassis, D, Bravo, J, Perisic, O, Pacold, C.M, Williams, R.L. | | Deposit date: | 2002-11-07 | | Release date: | 2002-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of the Px Domain of P47Phox to Phosphatidylinositol 3.4-Bisphosphate and Phosphatidic Acid is Masked by an Intramolecular Interaction
Embo J., 21, 2002
|
|
7SKU
 
 | | Nipah virus matrix protein in complex with PI(4,5)P2 | | Descriptor: | Matrix protein, SULFATE ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Norris, M.J, Saphire, E.O. | | Deposit date: | 2021-10-21 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Measles and Nipah virus assembly: Specific lipid binding drives matrix polymerization.
Sci Adv, 8, 2022
|
|
7SKS
 
 | |
7SKT
 
 | |
9CTW
 
 | |
9CTU
 
 | |
