1MZS
 
 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III WITH BOUND dichlorobenzyloxy-indole-carboxylic acid inhibitor | | Descriptor: | 1-(5-CARBOXYPENTYL)-5-(2,6-DICHLOROBENZYLOXY)-1H-INDOLE-2-CARBOXYLIC ACID, 3-oxoacyl-[acyl-carrier-protein] synthase III, PHOSPHATE ION | | Authors: | Daines, R.A, Pendrak, I, Sham, K, Van Aller, G.S, Konstantinidis, A.K, Lonsdale, J.T, Janson, C.A, Qui, X, Brandt, M, Silverman, C, Head, M.S. | | Deposit date: | 2002-10-09 | | Release date: | 2002-11-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | First X-ray cocrystal structure of a bacterial FabH condensing enzyme and a small molecule inhibitor achieved using rational design and homology modeling
J.Med.Chem., 46, 2003
|
|
5DNA
 
 | | Crystal structure of Candida boidinii formate dehydrogenase | | Descriptor: | FORMATE DEHYDROGENASE, SULFATE ION | | Authors: | Guo, Q, Gakhar, L, Wichersham, K, Francis, K, Vardi-Kilshtain, A, Major, D.T, Cheatum, C.M, Kohen, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Kinetic Studies of Formate Dehydrogenase from Candida boidinii.
Biochemistry, 55, 2016
|
|
5DN9
 
 | | Crystal structure of Candida boidinii formate dehydrogenase complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, FDH, ... | | Authors: | Guo, Q, Gakhar, L, Wichersham, K, Francis, K, Vardi-Kilshtain, A, Major, D.T, Cheatum, C.M, Kohen, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Kinetic Studies of Formate Dehydrogenase from Candida boidinii.
Biochemistry, 55, 2016
|
|
6CGW
 
 | |
6CHC
 
 | | JzTx-V toxin peptide, wild-type | | Descriptor: | Beta/kappa-theraphotoxin-Cg2a | | Authors: | Jordan, J.B. | | Deposit date: | 2018-02-22 | | Release date: | 2018-05-16 | | Method: | SOLUTION NMR | | Cite: | Pharmacological characterization of potent and selective NaV1.7 inhibitors engineered from Chilobrachys jingzhao tarantula venom peptide JzTx-V.
PLoS ONE, 13, 2018
|
|
6CNU
 
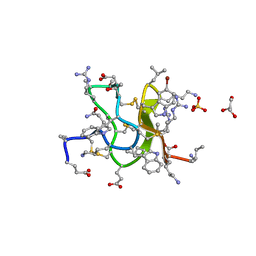 | | Crystal Structure of JzTX-V | | Descriptor: | BROMIDE ION, GLYCEROL, JzTx-V, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2018-03-09 | | Release date: | 2019-03-06 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Discovery of Tarantula Venom-Derived NaV1.7-Inhibitory JzTx-V Peptide 5-Br-Trp24 Analogue AM-6120 with Systemic Block of Histamine-Induced Pruritis.
J. Med. Chem., 61, 2018
|
|
3DS6
 
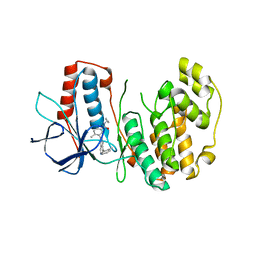 | | P38 complex with a phthalazine inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-[1-(2-methylphenyl)phthalazin-6-yl]benzamide | | Authors: | Herberich, B, Syed, R, Li, V, Grosfeld, D. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of highly selective and potent p38 inhibitors based on a phthalazine scaffold.
J.Med.Chem., 51, 2008
|
|
3DT1
 
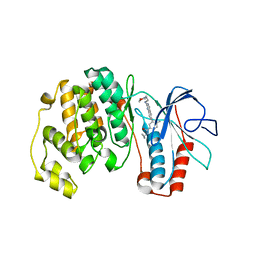 | | P38 Complexed with a quinazoline inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-{2-[(2-morpholin-4-ylethyl)amino]quinazolin-6-yl}benzamide | | Authors: | Herberich, B, Syed, R, Li, V, Tasker, A.S. | | Deposit date: | 2008-07-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of highly selective and potent p38 inhibitors based on a phthalazine scaffold.
J.Med.Chem., 51, 2008
|
|
6D4B
 
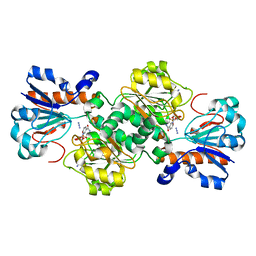 | | Crystal structure of Candida boidinii formate dehydrogenase V123A mutant complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Formate dehydrogenase, ... | | Authors: | Guo, Q, Ye, H, Gakhar, L, Cheatum, C.M, Kohen, A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oscillatory Active-site Motions Correlate with Kinetic Isotope Effects in Formate Dehydrogenase
Acs Catalysis, 2019
|
|
6D4C
 
 | | Crystal structure of Candida boidinii formate dehydrogenase V123G mutant complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Formate dehydrogenase, ... | | Authors: | Guo, Q, Ye, H, Gakhar, L, Cheatum, C.M, Kohen, A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oscillatory Active-site Motions Correlate with Kinetic Isotope Effects in Formate Dehydrogenase
Acs Catalysis, 2019
|
|
4BEB
 
 | | MUTANT (K220E) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4BEC
 
 | | MUTANT (K220A) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4BE7
 
 | | MUTANT (K220R) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
