5LIA
 
 | | Crystal structure of murine autotaxin in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Turnbull, A.P, Shah, P, Cheasty, A, Raynham, T, Pang, L, Owen, P. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of potent inhibitors of the lysophospholipase autotaxin.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
4GRV
 
 | | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Neurotensin 8-13, Neurotensin receptor type 1, ... | | Authors: | Noinaj, N, White, J.F, Shibata, Y, Love, J, Kloss, B, Xu, F, Gvozdenovic-Jeremic, J, Shah, P, Shiloach, J, Tate, C.G, Grisshammer, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the agonist-bound neurotensin receptor.
Nature, 490, 2012
|
|
4XES
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
4XEE
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
6Z43
 
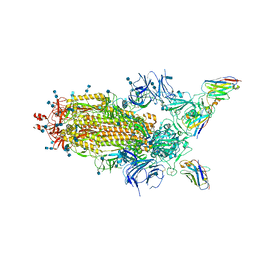 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
1ROV
 
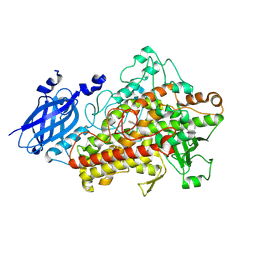 | | Lipoxygenase-3 Treated with Cumene Hydroperoxide | | Descriptor: | FE (III) ION, Seed lipoxygenase-3 | | Authors: | Vahedi-Faridi, A, Brault, P.A, Shah, P, Kim, Y.W, Dunham, W.R, Funk, M.O. | | Deposit date: | 2003-12-02 | | Release date: | 2004-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction between non-heme iron of lipoxygenases and cumene hydroperoxide: basis for enzyme activation, inactivation, and inhibition
J.Am.Chem.Soc., 126, 2004
|
|
1HV7
 
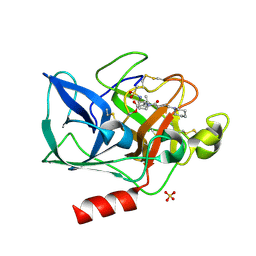 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH GW311616A | | Descriptor: | ELASTASE 1, N-[2-(1-FORMYL-2-METHYL-PROPYL)-1-(4-PIPERIDIN-1-YL-BUT-2-ENOYL)-PYRROLIDIN-3-YL]-METHANESULFONAMIDE, SULFATE ION | | Authors: | Skarzynski, T, Singh, O.M. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of a potent, intracellular, orally bioavailable, long duration inhibitor of human neutrophil elastase--GW311616A a development candidate
Bioorg.Med.Chem.Lett., 11, 2001
|
|
6E4A
 
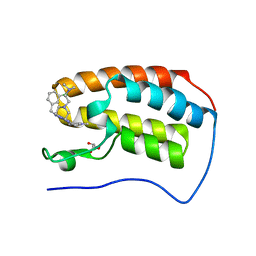 | | Crystal structure of human BRD4(1) in complex with CN750 | | Descriptor: | 5-(4-{[(3-chlorophenyl)methyl]amino}-2-{4-[2-(dimethylamino)ethyl]piperazin-1-yl}quinazolin-6-yl)-1-methylpyridin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Fontano, E, White, A, Lakshminarasimhan, D, Suto, R.K. | | Deposit date: | 2018-07-17 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery and lead identification of quinazoline-based BRD4 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7EH0
 
 | |
7EH2
 
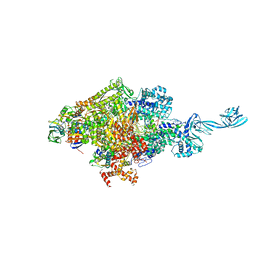 | |
7EH1
 
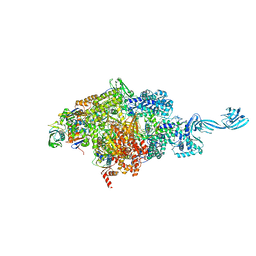 | | Thermus thermophilus transcription initiation complex containing a template-strand purine at position TSS-2, GpG RNA primer, and CMPcPP | | Descriptor: | 1,4-BUTANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (27-MER), ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Promoter-sequence determinants and structural basis of primer-dependent transcription initiation in Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Q0D
 
 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00384414-01 AT 2.05 A RESOLUTION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-[3-(cyclopentylethynyl)-4-fluorophenyl]-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Dranow, D.M, Davies, D.R. | | Deposit date: | 2019-08-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
6Q13
 
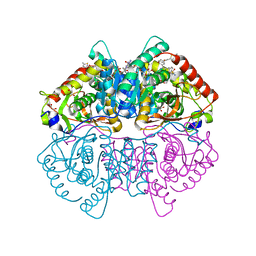 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00420737-09 AT 2.00 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-[5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-3-{3-[(5-methylthiophen-2-yl)ethynyl]phenyl}-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ... | | Authors: | Davies, D.R, Dranow, D.M. | | Deposit date: | 2019-08-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
1H1B
 
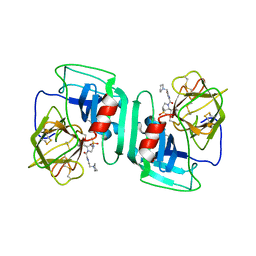 | | Crystal structure of human neutrophil elastase complexed with an inhibitor (GW475151) | | Descriptor: | (2S)-3-METHYL-2-((2R,3S)-3-[(METHYLSULFONYL)AMINO]-1-{[2-(PYRROLIDIN-1-YLMETHYL)-1,3-OXAZOL-4-YL]CARBONYL}PYRROLIDIN-2-YL)BUTANOIC ACID, LEUKOCYTE ELASTASE, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Macdonald, S.J.F, Dowle, M.D, Harrison, L.A, Clarke, G.D.E, Inglis, G.G.A, Johnson, M.R, Smith, R.A, Amour, A, Fleetwood, G, Humphreys, D.C, Molloy, C.R, Dixon, M, Godward, R.E, Wonacott, A.J, Singh, O.M.P, Hodgson, S.T, Hardy, G.W. | | Deposit date: | 2002-07-05 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Further Pyrrolidine Trans-Lactams as Inhibitors of Human Neutrophil Elastase (Hne) with Potential as Development Candidates and the Crystal Structure of Hne Complexed with an Inhibitor (Gw475151)
J.Med.Chem., 45, 2002
|
|
2JC0
 
 | | CRYSTAL STRUCTURE OF HEPATITIS C VIRUS POLYMERASE IN COMPLEX WITH INHIBITOR SB655264 | | Descriptor: | (2S,4S,5R)-2-ISOBUTYL-5-(2-THIENYL)-1-[4-(TRIFLUOROMETHYL)BENZOYL]PYRROLIDINE-2,4-DICARBOXYLIC ACID, RNA-DEPENDENT RNA-POLYMERASE | | Authors: | Wonacott, A, Skarzynski, T, Singh, O.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of Novel Acyl Pyrrolidine Inhibitors of Hepatitis C Virus RNA-Dependent RNA Polymerase Leading to a Development Candidate.
J.Med.Chem., 50, 2007
|
|
2JC1
 
 | | CRYSTAL STRUCTURE OF HEPATITIS C VIRUS POLYMERASE IN COMPLEX WITH INHIBITOR SB698223 | | Descriptor: | (2S,4S,5R)-1-(4-TERT-BUTYLBENZOYL)-2-ISOBUTYL-5-(1,3-THIAZOL-2-YL)PYRROLIDINE-2,4-DICARBOXYLIC ACID, RNA-DEPENDENT RNA-POLYMERASE | | Authors: | Wonacott, A, Skarzynski, T, Singh, O.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of Novel Acyl Pyrrolidine Inhibitors of Hepatitis C Virus RNA-Dependent RNA Polymerase Leading to a Development Candidate.
J.Med.Chem., 50, 2007
|
|
6DQB
 
 | | LINKED KDM5A JMJ DOMAIN FORMING COVALENT BOND TO INHIBITOR N71 i.e. 2-((3-(4-(dimethylamino)but-2-enamido)phenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 2-{(R)-(3-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(R)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(S)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ8
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR N49 i.e. 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, 2-[(S)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ6
 
 | |
6DQA
 
 | | Linked KDM5A JMJ Domain Bound to Inhibitor N70 i.e.[2-((3-aminophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-(3-aminophenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ4
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ9
 
 | | Linked KDM5A JMJ Domain Bound to the Covalent Inhibitor N69 i.e. [2-((3-acrylamidophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-[3-(acryloylamino)phenyl][2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ5
 
 | |
8PC7
 
 | |
6I8F
 
 | |
