4NTD
 
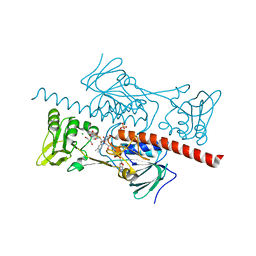 | | Crystal structure of HlmI | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NTC
 
 | | Crystal structure of GliT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GliT | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NTE
 
 | | Crystal structure of DepH | | Descriptor: | CHLORIDE ION, DepH, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4G91
 
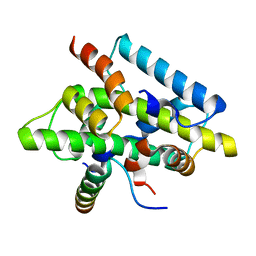 | | CCAAT-binding complex from Aspergillus nidulans | | Descriptor: | HAPB protein, HapE, Transcription factor HapC (Eurofung) | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
4G92
 
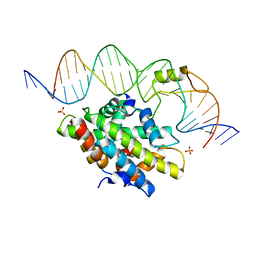 | | CCAAT-binding complex from Aspergillus nidulans with DNA | | Descriptor: | DNA, HAPB protein, HapE, ... | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
5LX1
 
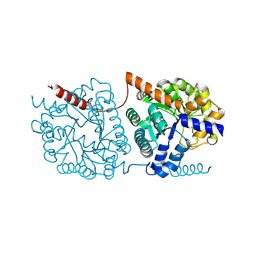 | | Cys-Gly dipeptidase GliJ mutant D304A | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LX7
 
 | | Cys-Gly dipeptidase GliJ mutant D38N | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dipeptidase, FE (III) ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LX0
 
 | | Cys-Gly dipeptidase GliJ (space group P3221) | | Descriptor: | Dipeptidase, FE (III) ION | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LX4
 
 | | Cys-Gly dipeptidase GliJ mutant D38H | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
7F58
 
 | | Cryo-EM structure of THIQ-MC4R-Gs_Nb35 complex | | Descriptor: | (3R)-N-[(2R)-3-(4-chlorophenyl)-1-[4-cyclohexyl-4-(1,2,4-triazol-1-ylmethyl)piperidin-1-yl]-1-oxidanylidene-propan-2-yl]-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F54
 
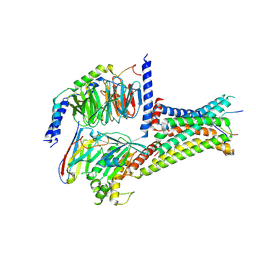 | | Cryo-EM structure of afamelanotide-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F55
 
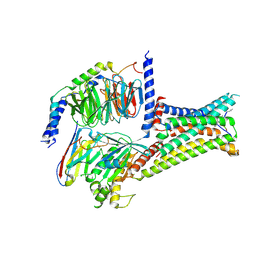 | | Cryo-EM structure of bremelanotide-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F53
 
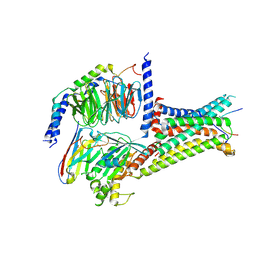 | | Cryo-EM structure of a-MSH-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
5NRY
 
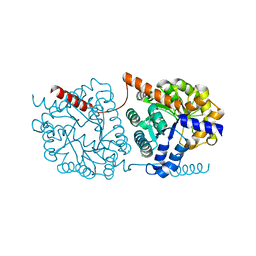 | | Cys-Gly dipeptidase GliJ in complex with Fe3+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, FE (III) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRX
 
 | | Cys-Gly dipeptidase GliJ in complex with Fe2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, FE (III) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS2
 
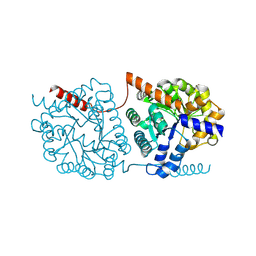 | | Cys-Gly dipeptidase GliJ in complex with Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Dipeptidase gliJ, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS1
 
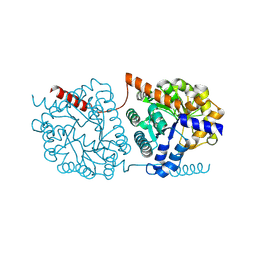 | | Cys-Gly dipeptidase GliJ in complex with Ni2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, NICKEL (II) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRZ
 
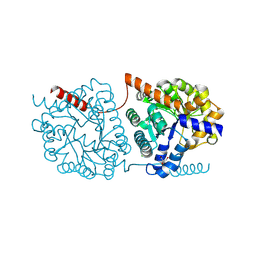 | | Cys-Gly dipeptidase GliJ in complex with Mn2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, GLYCEROL, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LWZ
 
 | | Cys-Gly dipeptidase GliJ (space group C2) | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRT
 
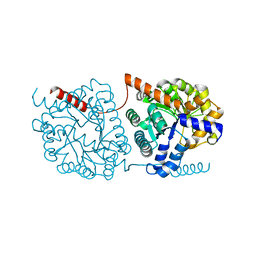 | | Cys-Gly dipeptidase GliJ in complex with Ca2+ | | Descriptor: | CALCIUM ION, Dipeptidase gliJ, MAGNESIUM ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS5
 
 | | Cys-Gly dipeptidase GliJ in complex with Cu2+ and Zn2+ | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Dipeptidase gliJ, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRU
 
 | | Cys-Gly dipeptidase GliJ in complex with Zn2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, ZINC ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
7NCB
 
 | | Glutathione-S-transferase GliG mutant H26A | | Descriptor: | Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NC9
 
 | | Glutathione-S-transferase GliG mutant H26N | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NCM
 
 | | Glutathione-S-transferase GliG mutant E82A | | Descriptor: | 1,2-ETHANEDIOL, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
