2BHN
 
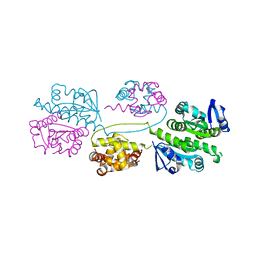 | | XPF from Aeropyrum pernix | | Descriptor: | XPF ENDONUCLEASE | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
2BGW
 
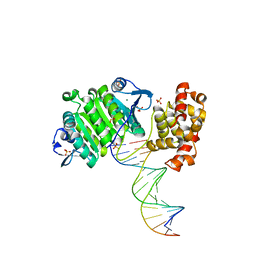 | | XPF from Aeropyrum pernix, complex with DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*AP*CP*AP*GP*AP*TP *GP*CP*TP*GP*A)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*GP *TP*GP*AP*TP*C)-3', MAGNESIUM ION, ... | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4Z5Q
 
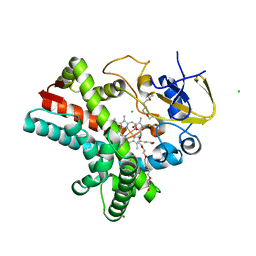 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
5BMO
 
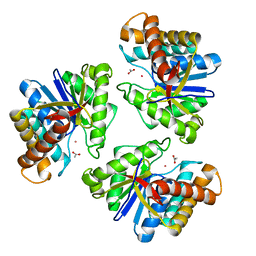 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
5F1P
 
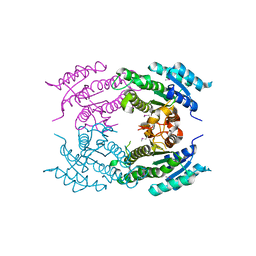 | | Crystal Structure of Dehydrogenase from Streptomyces platensis | | Descriptor: | PtmO8 | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.-Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-11-30 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal Structure of a Dehydrogenase, PtmO8, from Streptomyces platensis
To Be Published
|
|
4ZNM
 
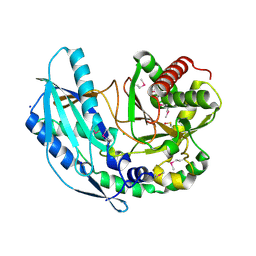 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | Descriptor: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
4ZXW
 
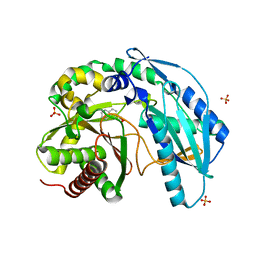 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | Descriptor: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
4AFV
 
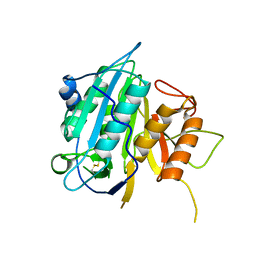 | | THE STRUCTURE OF METACASPASE 2 FROM T. BRUCEI DETERMINED IN THE PRESENCE OF CALCIUM CHLORIDE | | Descriptor: | METACASPASE MCA2 | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AFR
 
 | | THE STRUCTURE OF METACASPASE 2 (C213A MUTANT) FROM T. BRUCEI | | Descriptor: | METACASPASE | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AFP
 
 | | The structure of metacaspase 2 from T. brucei determined in the presence of Samarium | | Descriptor: | METACASPASE MCA2, SAMARIUM (III) ION | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AF8
 
 | | The structural basis for metacaspase substrate specificity and activation | | Descriptor: | CHLORIDE ION, METACASPASE MCA2, SODIUM ION | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-18 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5E7Q
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis | | Descriptor: | GLYCEROL, SULFATE ION, acyl-CoA synthetase | | Authors: | Osipiuk, J, Cuff, M.E, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-12 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
2VL7
 
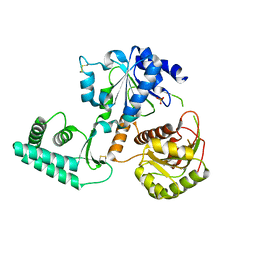 | | Structure of S. tokodaii Xpd4 | | Descriptor: | PHOSPHATE ION, XPD | | Authors: | Naismith, J.H, Johnson, K.A, Oke, M, McMahon, S.A, Liu, L, White, M.F, Zawadski, M, Carter, L.G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the DNA Repair Helicase Xpd.
Cell(Cambridge,Mass.), 133, 2008
|
|
2IX2
 
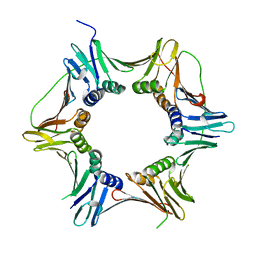 | | Crystal structure of the heterotrimeric PCNA from Sulfolobus solfataricus | | Descriptor: | DNA POLYMERASE SLIDING CLAMP A, DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C | | Authors: | Williams, G.J, Johnson, K, McMahon, S.A, Carter, L, Oke, M, Liu, H, Taylor, G.L, White, M.F, Naismith, J.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Heterotrimeric PCNA from Sulfolobus Solfataricus.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
