6M65
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPNP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M69
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPCP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M6Y
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M72
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGDP | | Descriptor: | 2'-deoxy-8-oxoguanosine 5'-(trihydrogen diphosphate), Hydrolase, NUDIX family protein, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2N0K
 
 | | Chemical shift assignments and structure of the alpha-crystallin domain from human, HSPB5 | | Descriptor: | Alpha-crystallin B chain | | Authors: | Rajagopal, P, Klevit, R.E, Shi, L, Baker, D. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis.
Elife, 4, 2015
|
|
6BP9
 
 | |
2N3J
 
 | | Solution Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1 | | Descriptor: | Heat shock protein beta-1 | | Authors: | Rajagopal, P, Liu, Y, Shi, L, Klevit, R.E. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1.
J.Biomol.Nmr, 63, 2015
|
|
8I61
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I67
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 2,4-Thiazolidinedione, Form I | | Descriptor: | 1,2-ETHANEDIOL, 1,3-thiazolidine-2,4-dione, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I69
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Fluoroorotic acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6B
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Hydroxy-2,4(1H,3H)-pyrimidinedione, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-oxidanyl-1~{H}-pyrimidine-2,4-dione, CHLORIDE ION, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I63
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I66
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with isoorotic acid (2,4-Dihydroxypyrimidine-5-carboxylic Acid) and citric acid, Form I | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, CITRIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I62
 
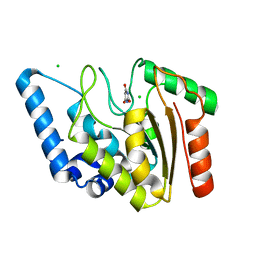 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, CHLORIDE ION, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I65
 
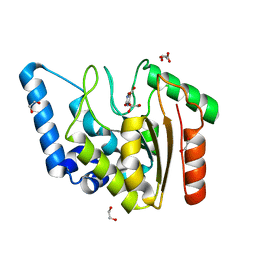 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with isoorotic acid (2,4-Dihydroxypyrimidine-5-carboxylic Acid), Form I | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6A
 
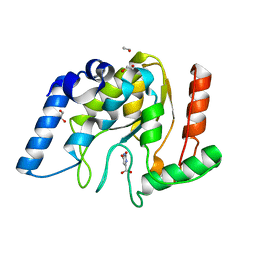 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Orotic acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, OROTIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6C
 
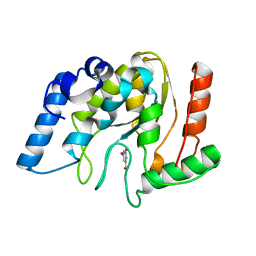 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 6-Formyl-uracil, Form III | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I64
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form II | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I68
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Uric acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, URIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6D
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Hydroxy-2,4(1H,3H)-pyrimidinedione, Form VI | | Descriptor: | 1,2-ETHANEDIOL, 5-oxidanyl-1~{H}-pyrimidine-2,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
9EY4
 
 | | The FK1 domain of FKBP51 in complex with (3S,11S)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxamide | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding Proteins.
Chemistry, 30, 2024
|
|
9EY3
 
 | | The FK1 domain of FKBP51 in complex with (3S,11S,11aS)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxylic acid | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxylic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding Proteins.
Chemistry, 30, 2024
|
|
1NOV
 
 | | NODAMURA VIRUS | | Descriptor: | NODAMURA VIRUS COAT PROTEINS | | Authors: | Natarajan, P, Johnson, J.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Resolution of space-group ambiguity and structure determination of nodamura virus to 3.3 A resolution from pseudo-R32 (monoclinic) crystals.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1JEM
 
 | | NMR STRUCTURE OF HISTIDINE PHOSPHORYLATED FORM OF THE PHOSPHOCARRIER HISTIDINE CONTAINING PROTEIN FROM BACILLUS SUBTILIS, NMR, 25 STRUCTURES | | Descriptor: | HISTIDINE CONTAINING PROTEIN | | Authors: | Jones, B.E, Rajagopal, P, Klevit, R.E. | | Deposit date: | 1997-04-01 | | Release date: | 1997-07-23 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation on histidine is accompanied by localized structural changes in the phosphocarrier protein, HPr from Bacillus subtilis.
Protein Sci., 6, 1997
|
|
3J07
 
 | | Model of a 24mer alphaB-crystallin multimer | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | Deposit date: | 2011-04-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | Cite: | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
