8TMT
 
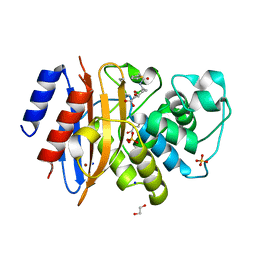 | | Crystal structure of KPC-44 carbapenemase in complex with vaborbactam | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, LITHIUM ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Neetu, N, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TN0
 
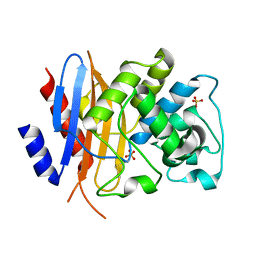 | | Crystal structure of KPC-44 carbapenemase w/o cryoprotectant | | Descriptor: | SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TJM
 
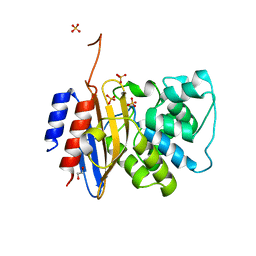 | | Crystal structure of KPC-44 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, beta-lactamase | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TMR
 
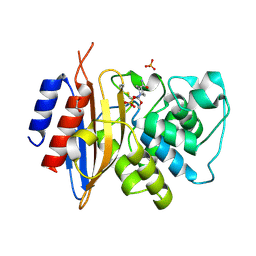 | | Crystal structure of KPC-44 carbapenemase complexed with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
4S2L
 
 | | Crystal Structure of OXA-163 beta-lactamase | | Descriptor: | Beta-lactamase, SODIUM ION | | Authors: | Stojanoski, V, Liya, H, Palzkill, T.G, Prasad, B, Sankaran, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
4S2M
 
 | | Crystal Structure of OXA-163 complexed with iodide in the active site | | Descriptor: | Beta-lactamase, IODIDE ION | | Authors: | Stojanoski, V, Hu, L, Palzkill, T.G, Prasad, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
7JIE
 
 | |
7MRY
 
 | | Norovirus T=3 GII.4 HOV VLP | | Descriptor: | VP1 | | Authors: | Salmen, W, Hu, L, Prasad, B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the predominant GII.4 human norovirus capsid reveals novel stability and plasticity.
Nat Commun, 13, 2022
|
|
8R7V
 
 | | MutSbeta bound to compound CHDI-00898647 in the canonical DNA-mismatch bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Haque, T, Brace, G, Felsenfeld, D, Tilack, K, Schaertl, S, Ballantyne, G, Maillard, M, Iyer, R, Prasad, B, Thomsen, M, Wilkionson, H, Plotnikov, N, Ritzefeld, M. | | Deposit date: | 2023-11-27 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Identification of orthosteric inhibitors of MutSbeta ATPase function
To Be Published
|
|
