5H31
 
 | |
5H33
 
 | |
5VJI
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
5VJX
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
1Y9J
 
 | | Solution structure of the rat Sly1 N-terminal domain | | Descriptor: | Sec1 family domain containing protein 1 | | Authors: | Arac, D, Dulubova, I, Pei, J, Huryeva, I, Grishin, N.V, Rizo, J. | | Deposit date: | 2004-12-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Structure of the rSly1 N-terminal Domain Reveals a Conformational Change Induced by Binding to Syntaxin 5.
J.Mol.Biol., 346, 2005
|
|
3TKK
 
 | | Crystal Structure Analysis of a recombinant predicted acetamidase/ formamidase from the thermophile thermoanaerobacter tengcongensis | | Descriptor: | CALCIUM ION, Predicted acetamidase/formamidase, ZINC ION | | Authors: | Qian, M, Huang, Q, Wu, G, Lai, L, Tang, Y, Pei, J, Kusunoki, M. | | Deposit date: | 2011-08-26 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure Analysis of a Recombinant Predicted Acetamidase/Formamidase from the Thermophile Thermoanaerobacter tengcongensis.
PROTEIN J., 31, 2012
|
|
1DU9
 
 | | SOLUTION STRUCTURE OF BMP02, A NATURAL SCORPION TOXIN WHICH BLOCKS APAMIN-SENSITIVE CALCIUM-ACTIVATED POTASSIUM CHANNELS, 25 STRUCTURES | | Descriptor: | BMP02 NEUROTOXIN | | Authors: | Xu, Y, Wu, J, Pei, J, Shi, Y, Ji, Y, Tong, Q. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-04 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP02, a new potassium channel blocker from the venom of the Chinese scorpion Buthus martensi Karsch.
Biochemistry, 39, 2000
|
|
7LDK
 
 | | Structure of human respiratory syncytial virus nonstructural protein 2 (NS2) | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, Non-structural protein 2 | | Authors: | Chatterjee, S, Borek, D, Otwinowski, Z, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2021-01-13 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for IFN antagonism by human respiratory syncytial virus nonstructural protein 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7P
 
 | | Cryo-EM structure of human TMEM120A | | Descriptor: | COENZYME A, Ion channel TACAN | | Authors: | Xue, J, Han, Y, Jiang, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | TMEM120A is a coenzyme A-binding membrane protein with structural similarities to ELOVL fatty acid elongase.
Elife, 10, 2021
|
|
7LIP
 
 | | X-ray structure of SPOP MATH domain (D140G) | | Descriptor: | SULFATE ION, Speckle-type POZ protein | | Authors: | Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | ATM-phosphorylated SPOP contributes to 53BP1 exclusion from chromatin during DNA replication.
Sci Adv, 7, 2021
|
|
7LIO
 
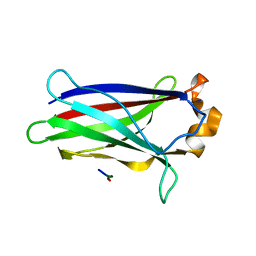 | |
7LIQ
 
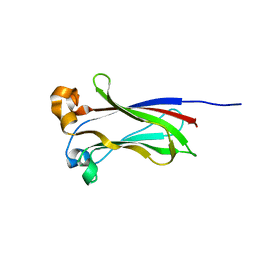 | |
7LIN
 
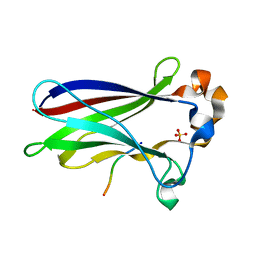 | |
5MF1
 
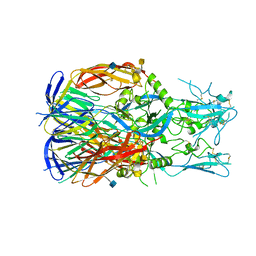 | |
5IT3
 
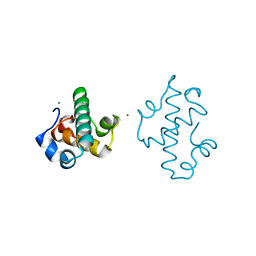 | | Swirm domain of human Lsd1 | | Descriptor: | Lysine-specific histone demethylase 1A, MAGNESIUM ION | | Authors: | Jeffrey, P.D, Yuan, P. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Tlx-interacting peptide of Lsd1 inhibits the proliferation of brain tumor stem cells
To Be Published
|
|
4ZBW
 
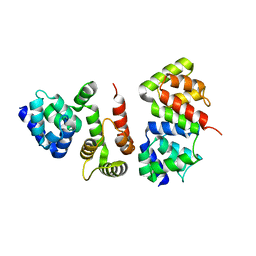 | |
3WGH
 
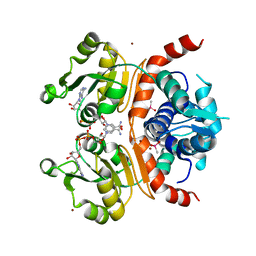 | | Crystal structure of RSP in complex with beta-NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CACODYLATE ION, Redox-sensing transcriptional repressor rex, ... | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
3WGI
 
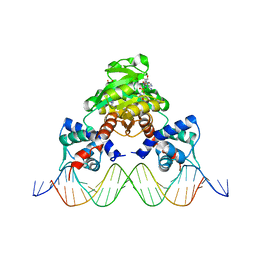 | | Crystal structure of RSP in complex with beta-NAD+ and operator DNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*TP*GP*TP*TP*AP*AP*TP*CP*GP*AP*TP*TP*AP*AP*CP*AP*AP*TP*C)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), Redox-sensing transcriptional repressor rex | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
3WG9
 
 | | Crystal structure of RSP, a Rex-family repressor | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
1JAZ
 
 | |
1JJA
 
 | |
1IHD
 
 | |
8VTT
 
 | | Meis1 homeobox domain bound to neomycin fragment | | Descriptor: | Homeobox protein Meis1, RIBOSTAMYCIN, SULFATE ION | | Authors: | Tomchick, D.R, Ahmed, M.S, Nguyen, N.U.N, Sadek, H.A. | | Deposit date: | 2024-01-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of FDA-approved drugs that induce heart regeneration in mammals.
Nat Cardiovasc Res, 3, 2024
|
|
8VTS
 
 | | Meis1 homeobox domain bound to paromomycin fragment | | Descriptor: | 1,2-ETHANEDIOL, Homeobox protein Meis1, ISOPROPYL ALCOHOL, ... | | Authors: | Tomchick, D.R, Ahmed, M.S, Nguyen, N.U.N. | | Deposit date: | 2024-01-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of FDA-approved drugs that induce heart regeneration in mammals.
Nat Cardiovasc Res, 3, 2024
|
|
3E8K
 
 | |
