5ED9
 
 | | Crystal structure of CC1 of mouse SUN2 | | Descriptor: | SUN domain-containing protein 2 | | Authors: | Nie, S, Ke, H.M, Gao, F, Ren, J.Q, Wang, M.Z, Huo, L, Gong, W.M, Feng, W. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators
Structure, 24, 2016
|
|
5ED8
 
 | | Crystal structure of CC2-SUN of mouse SUN2 | | Descriptor: | MAGNESIUM ION, MKIAA0668 protein | | Authors: | Nie, S, Ke, H.M, Gao, F, Ren, J.Q, Wang, M.Z, Huo, L, Gong, W.M, Feng, W. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators
Structure, 24, 2016
|
|
1PP2
 
 | |
1N1C
 
 | | Crystal Structure Of The Dimeric TorD Chaperone From Shewanella Massilia | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TorA specific chaperone | | Authors: | Tranier, S, Iobbi-Nivol, C, Mortier-Barriere, I, Birck, C, Mejean, V, Samama, J.-P. | | Deposit date: | 2002-10-17 | | Release date: | 2003-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel Protein Fold and Extreme Domain Swapping in the Dimeric TorD Chaperone from Shewanella massilia
Structure, 11, 2003
|
|
1D1N
 
 | | SOLUTION STRUCTURE OF THE FMET-TRNAFMET BINDING DOMAIN OF BECILLUS STEAROTHERMOPHILLUS TRANSLATION INITIATION FACTOR IF2 | | Descriptor: | INITIATION FACTOR 2 | | Authors: | Meunier, S, Spurio, S, Czisch, M, Wechselberger, R, Gueunneugues, M. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the fMet-tRNA(fMet)-binding domain of B. stearothermophilus initiation factor IF2.
EMBO J., 19, 2000
|
|
2KPK
 
 | | MAGI-1 PDZ1 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
6S45
 
 | | Room temperature structure of the dark state of the LOV2 domain of phototropin-2 from Arabidopsis thaliana determined with a serial crystallography approach | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Aumonier, S, Santoni, G, Gotthard, G, von Stetten, D, Leonard, G, Royant, A. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Millisecond time-resolved serial oscillation crystallography of a blue-light photoreceptor at a synchrotron.
Iucrj, 7, 2020
|
|
1E25
 
 | | The high resolution structure of PER-1 class A beta-lactamase | | Descriptor: | EXTENDED-SPECTRUM BETA-LACTAMASE PER-1, SULFATE ION | | Authors: | Tranier, S, Bouthors, A.T, Maveyraud, L, Guillet, V, Sougakoff, W, Samama, J.P. | | Deposit date: | 2000-05-17 | | Release date: | 2000-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The High Resolution Crystal Structure for Class a Beta-Lactamase Per-1 Reveals the Bases for its Increase in Breadth of Activity
J.Biol.Chem., 275, 2000
|
|
1NCV
 
 | | DETERMINATION CC-CHEMOKINE MCP-3, NMR, 7 STRUCTURES | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 3 | | Authors: | Meunier, S, Bernassau, J.M, Guillemot, J.C, Ferrara, P, Darbon, H. | | Deposit date: | 1997-02-05 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of CC chemokine monocyte chemoattractant protein 3 by 1H two-dimensional NMR spectroscopy.
Biochemistry, 36, 1997
|
|
6S46
 
 | | Room temperature structure of the LOV2 domain of phototropin-2 from Arabidopsis thaliana 4158 ms after initiation of illumination, determined with a serial crystallography approach | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Aumonier, S, Santoni, G, Gotthard, G, von Stetten, D, Leonard, G, Royant, A. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Millisecond time-resolved serial oscillation crystallography of a blue-light photoreceptor at a synchrotron.
Iucrj, 7, 2020
|
|
1PNH
 
 | | SOLUTION STRUCTURE OF PO5-NH2, A SCORPION TOXIN ANALOG WITH HIGH AFFINITY FOR THE APAMIN-SENSITIVE POTASSIUM CHANNEL | | Descriptor: | SCORPION TOXIN | | Authors: | Meunier, S, Bernassau, J.-M, Sabatier, J.-M, Martin-Eauclaire, M.-F, Van Rietschoten, J, Cambillau, C, Darbon, H. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P05-NH2, a scorpion toxin analog with high affinity for the apamin-sensitive potassium channel.
Biochemistry, 32, 1993
|
|
4EJ4
 
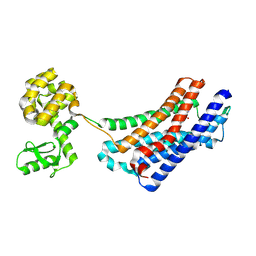 | | Structure of the delta opioid receptor bound to naltrindole | | Descriptor: | (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,14,14b-hexahydro-4,8-methano[1]benzofuro[2,3-a]pyrido[4,3-b]carbazole-1,8a(9H)-diol, Delta-type opioid receptor, Lysozyme chimera | | Authors: | Granier, S, Manglik, A, Kruse, A.C, Kobilka, T.S, Thian, F.S, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of the delta opioid receptor bound to naltrindole
Nature, 485, 2012
|
|
1RJH
 
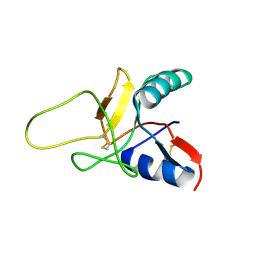 | | Structure of the Calcium Free Form of the C-type Lectin-like Domain of Tetranectin | | Descriptor: | Tetranectin | | Authors: | Nielbo, S, Thomsen, J.K, Graversen, J.H, Etzerodt, M, Poulsen, F.M, Thoegersen, H.C. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the Plasminogen Kringle 4 Binding Calcium-Free Form of the C-Type Lectin-Like Domain of Tetranectin.
Biochemistry, 43, 2004
|
|
2KPL
 
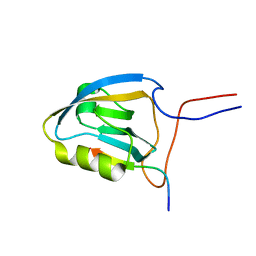 | | MAGI-1 PDZ1 / E6CT | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, Protein E6 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
6QQK
 
 | |
6QQI
 
 | |
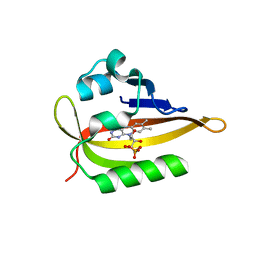
6QQJ
 
 | |
6QQH
 
 | |
6QSA
 
 | |
5LK4
 
 | | Structure of the Red Fluorescent Protein mScarlet at pH 7.8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Aumonier, S, Gotthard, G, Royant, A. | | Deposit date: | 2016-07-20 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | mScarlet: a bright monomeric red fluorescent protein for cellular imaging.
Nat. Methods, 14, 2017
|
|
6MO1
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 5-[4-(aminomethyl)phenyl]-6-[4-(furan-3-yl)phenyl]-N-[(piperidin-4-yl)methyl]pyrazin-2-amine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Hua, Y, Nie, S, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
6MO0
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{3-[4-(furan-3-yl)phenyl]-5-[(piperidin-4-yl)methoxy]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J. Am. Chem. Soc., 141, 2019
|
|
6MO2
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{5-[(piperidin-4-yl)methoxy]-3-[4-(1H-pyrazol-4-yl)phenyl]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
7TBI
 
 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | Descriptor: | Dyn2, Nic96 R1, Nic96 R2, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBM
 
 | | Composite structure of the dilated human nuclear pore complex (NPC) generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | Descriptor: | DDX19, NUP107 CTD, NUP107 NTD, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
