6BSR
 
 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6BSQ
 
 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | Descriptor: | CHLORIDE ION, GLYCEROL, PBP4 protein | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKA
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the open conformation | | Descriptor: | SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKG
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the benzylpenicilin-bound form | | Descriptor: | OPEN FORM - PENICILLIN G, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKJ
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the closed conformation | | Descriptor: | penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Soares, A, D'Andrea, E.D, Jaconcic, J, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
2LRU
 
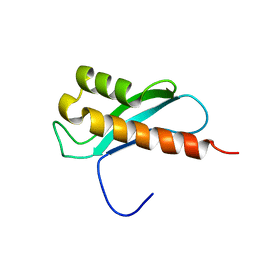 | | Solution Structure of the WNK1 Autoinhibitory Domain | | Descriptor: | Serine/threonine-protein kinase WNK1 | | Authors: | Moon, T.M, Correa, F, Gardner, K.H, Goldsmith, E.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the WNK1 Autoinhibitory Domain, a WNK-Specific PF2 Domain.
J.Mol.Biol., 425, 2013
|
|
6MKH
 
 | | Crystal structure of pencillin binding protein 4 (PBP4) from Enterococcus faecalis in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PHOSPHATE ION, pencillin binding protein 4 (PBP4) | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKI
 
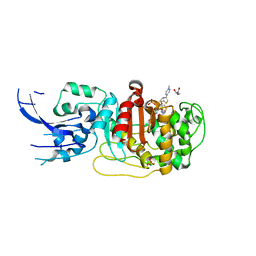 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the ceftaroline-bound form | | Descriptor: | Ceftaroline, bound form, GLYCEROL, ... | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6OBR
 
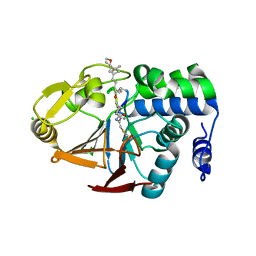 | | PP1 Y134A in complex with Microcystin LR | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Microcystin LR, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBS
 
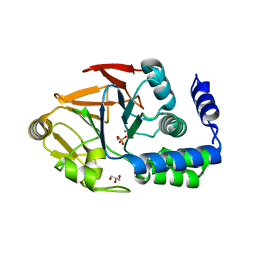 | | PP1 Y134K | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBN
 
 | | The crystal structure of coexpressed SDS22:PP1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBQ
 
 | | PP1 H66K in complex with Microcystin LR | | Descriptor: | MANGANESE (II) ION, Microcystin LR, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBP
 
 | | Reconstituted PP1 holoenzyme | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBU
 
 | | PP1 Y134K in complex with Microcystin LR | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4PWN
 
 | | Crystal structure of Active WNK1 kinase | | Descriptor: | PHOSPHATE ION, Serine/threonine-protein kinase WNK1 | | Authors: | Piala, A, Moon, T, Akella, T, He, H, Cobbm, M.H, Goldsmith, E. | | Deposit date: | 2014-03-20 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Chloride Sensing by WNK1 Involves Inhibition of Autophosphorylation.
Sci.Signal., 7, 2014
|
|
4Q2A
 
 | | WNK1: A chloride sensor via autophosphorylation | | Descriptor: | BROMIDE ION, Serine/threonine-protein kinase WNK1 | | Authors: | Piala, A, Moon, T, Akella, R, He, H, Cobb, M.H, Goldsmith, E. | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Chloride Sensing by WNK1 Involves Inhibition of Autophosphorylation.
Sci.Signal., 7, 2014
|
|
