1YMG
 
 | | The Channel Architecture of Aquaporin O at 2.2 Angstrom Resolution | | Descriptor: | Lens fiber major intrinsic protein, nonyl beta-D-glucopyranoside | | Authors: | Harries, W.E.C, Akhavan, D, Miercke, L.J.W, Khademi, S, Stroud, R.M. | | Deposit date: | 2005-01-20 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Channel Architecture of Aquaporin 0 at a 2.2-A Resolution
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U7C
 
 | | Crystal Structure of AmtB from E.Coli with Methyl Ammonium. | | Descriptor: | METHYLAMINE, Probable ammonium transporter | | Authors: | Khademi, S, O'Connell III, J, Remis, J, Robles-Colmenares, Y, Miercke, L.J.W, Stroud, R.M. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A
Science, 305, 2004
|
|
1U77
 
 | | Crystal Structure of Ammonia Channel AmtB from E. Coli | | Descriptor: | Probable ammonium transporter | | Authors: | Khademi, S, O'Connell III, J, Remis, J, Robles-Colmenares, Y, Miercke, L.J.W, Stroud, R.M. | | Deposit date: | 2004-08-02 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A
Science, 305, 2004
|
|
1U7G
 
 | | Crystal Structure of Ammonia Channel AmtB from E. Coli | | Descriptor: | AMMONIA, AMMONIUM ION, Probable ammonium transporter, ... | | Authors: | Khademi, S, O'Connell III, J, Remis, J, Robles-Colmenares, Y, Miercke, L.J.W, Stroud, R.M. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A
Science, 305, 2004
|
|
1NM2
 
 | | Malonyl-CoA:ACP Transacylase | | Descriptor: | ACETIC ACID, NICKEL (II) ION, malonyl CoA:acyl carrier protein malonyltransferase | | Authors: | Keatinge-Clay, A.T, Shelat, A.A, Savage, D.F, Tsai, S, Miercke, L.J.W, O'Connell III, J.D, Khosla, C, Stroud, R.M. | | Deposit date: | 2003-01-08 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalysis, Specificity, and ACP Docking Site of
Streptomyces coelicolor Malonyl-CoA:ACP Transacylase
Structure, 11, 2003
|
|
1KEZ
 
 | | Crystal Structure of the Macrocycle-forming Thioesterase Domain of Erythromycin Polyketide Synthase (DEBS TE) | | Descriptor: | ERYTHRONOLIDE SYNTHASE | | Authors: | Tsai, S.-C, Miercke, L.J.W, Krucinski, J, Gokhale, R, Chen, J.C.-H, Foster, P.G, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4HB1
 
 | | A DESIGNED FOUR HELIX BUNDLE PROTEIN. | | Descriptor: | DHP1, UNKNOWN ATOM OR ION | | Authors: | Schafmeister, C.E, Laporte, S.L, Miercke, L.J.W, Stroud, R.M. | | Deposit date: | 1997-11-10 | | Release date: | 1998-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A designed four helix bundle protein with native-like structure.
Nat.Struct.Biol., 4, 1997
|
|
1FX8
 
 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITH SUBSTRATE GLYCEROL | | Descriptor: | GLYCEROL, GLYCEROL UPTAKE FACILITATOR PROTEIN, octyl beta-D-glucopyranoside | | Authors: | Fu, D, Libson, A, Miercke, L.J.W, Weitzman, C, Nollert, P, Stroud, R.M. | | Deposit date: | 2000-09-25 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a glycerol-conducting channel and the basis for its selectivity.
Science, 290, 2000
|
|
3GD8
 
 | | Crystal Structure of Human Aquaporin 4 at 1.8 and its Mechanism of Conductance | | Descriptor: | Aquaporin-4, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Ho, J.D, Yeh, R, Sandstrom, A, Chorny, I, Harries, W.E.C, Robbins, R.A, Miercke, L.J.W, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1EXQ
 
 | | CRYSTAL STRUCTURE OF THE HIV-1 INTEGRASE CATALYTIC CORE DOMAIN | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-05-03 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EX4
 
 | | HIV-1 INTEGRASE CATALYTIC CORE AND C-TERMINAL DOMAIN | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, INTEGRASE | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2BCE
 
 | | CHOLESTEROL ESTERASE FROM BOS TAURUS | | Descriptor: | CHOLESTEROL ESTERASE | | Authors: | Chen, J.C.-H, Miercke, L.J.W, Krucinski, J, Starr, J.R, Saenz, G, Wang, X, Spilburg, C.A, Lange, L.G, Ellsworth, J.L, Stroud, R.M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic cholesterol esterase at 1.6 A: novel structural features involved in lipase activation.
Biochemistry, 37, 1998
|
|
1LDA
 
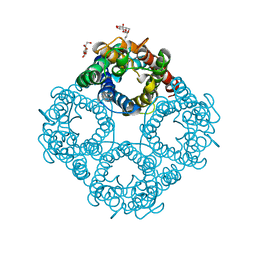 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LDI
 
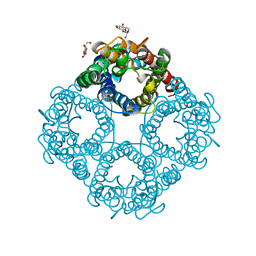 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LDF
 
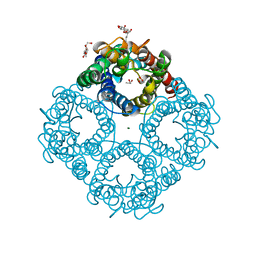 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) MUTATION W48F, F200T | | Descriptor: | GLYCEROL, Glycerol uptake facilitator protein, MAGNESIUM ION, ... | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1MZJ
 
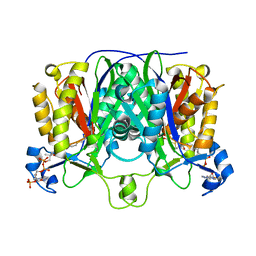 | | Crystal Structure of the Priming beta-Ketosynthase from the R1128 Polyketide Biosynthetic Pathway | | Descriptor: | ACETYL GROUP, Beta-ketoacylsynthase III, COENZYME A | | Authors: | Pan, H, Tsai, S.C, Meadows, E.S, Miercke, L.J.W, Keatinge-Clay, A, O'Connell, J, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-10-08 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the priming beta-ketosynthase from the
R1128 polyketide biosynthetic pathway
Structure, 10
|
|
1NN6
 
 | | Human Pro-Chymase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase | | Authors: | Reiling, K.K, Krucinski, J, Miercke, L.J.W, Raymond, W.W, Caughey, G.H, Stroud, R.M. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human pro-chymase: a model for the activating transition of granule-associated proteases.
Biochemistry, 42, 2003
|
|
2B6P
 
 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | Descriptor: | Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
