2X2B
 
 | | Crystal structure of malonyl-ACP (acyl carrier protein) from Bacillus subtilis | | Descriptor: | 3-{[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-3-oxopropanoic acid, ACYL CARRIER PROTEIN, PLATINUM (II) ION | | Authors: | Bellinzoni, M, Martinez, M.A, DeMendoza, D, Alzari, P.M. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Novel Role of Malonyl-Acp in Lipid Homeostasis.
Biochemistry, 49, 2010
|
|
2XKG
 
 | | C.lacteus mini-Hb Leu86Ala mutant | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2XKH
 
 | | Xe derivative of C.lacteus mini-Hb Leu86Ala mutant | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2XKI
 
 | | Aquo-met structure of C.lacteus mini-Hb | | Descriptor: | GLYCEROL, NEURAL HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
6PQ1
 
 | | Structure of the Fremyella diplosiphon OCP1 | | Descriptor: | Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Dominguez-Martin, M.A, Bao, H, Kerfeld, C.A. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Comparative ultrafast spectroscopy and structural analysis of OCP1 and OCP2 from Tolypothrix.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
6MCJ
 
 | |
7SCB
 
 | | B-cylinder of Synechocystis PCC 6803 Phycobilisome, complex with OCP - local refinement | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SCA
 
 | | Synechocystis PCC 6803 Phycobilisome rod from OCP-PBS complex sample | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SCC
 
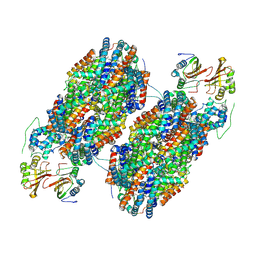 | | T-cylinder of Synechocystis PCC 6803 Phycobilisome, complex with OCP - local refinement | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Orange carotenoid-binding protein, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SC8
 
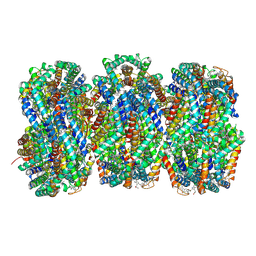 | | Synechocystis PCC 6803 Phycobilisome rod from PBS sample | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SC7
 
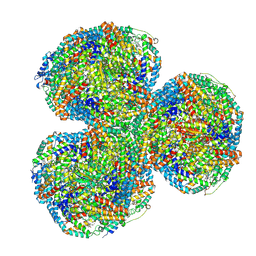 | | Synechocystis PCC 6803 Phycobilisome core from up-down rod conformation | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SC9
 
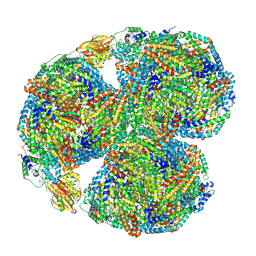 | | Synechocystis PCC 6803 Phycobilisome core, complex with OCP | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
9FM4
 
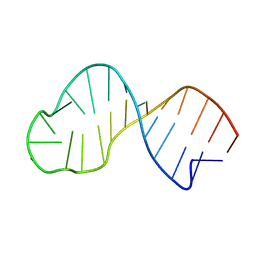 | | Dynamic structure of the apical stem loop of the stem loop 2 motif (s2m) from SCoV-2 Delta variant | | Descriptor: | RNA (25-MER) | | Authors: | Wirtz Martin, M.A, Matzel, T, Makowski, J, Kensinger, A, Herr, A, Wacker, A, Richter, C, Jonker, H.R.A, Evanseck, J, Schwalbe, H. | | Deposit date: | 2024-06-05 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-16 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structural heterogeneity and dynamics in the apical stem loop of s2m from SARS-CoV-2 Delta by an integrative NMR spectroscopy and MD simulation approach.
Nucleic Acids Res., 53, 2025
|
|
8BJ8
 
 | |
8BJ7
 
 | |
5IUK
 
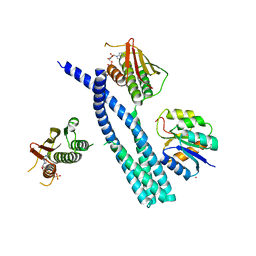 | | Crystal structure of the DesK-DesR complex in the phosphotransfer state with high Mg2+ (150 mM) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Trajtenberg, F, Imelio, J.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2016-03-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action.
Elife, 5, 2016
|
|
5IUM
 
 | | Crystal structure of phosphorylated DesKC | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Trajtenberg, F, Buschiazzo, A. | | Deposit date: | 2016-03-18 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action.
Elife, 5, 2016
|
|
5IUL
 
 | | Crystal structure of the DesK-DesR complex in the phosphotransfer state with high Mg2+ (150 mM) and BeF3 | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Trajtenberg, F, Imelio, J.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2016-03-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action.
Elife, 5, 2016
|
|
5IUJ
 
 | | Crystal structure of the DesK-DesR complex in the phosphotransfer state with low Mg2+ (20 mM) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Trajtenberg, F, Imelio, J.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2016-03-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action.
Elife, 5, 2016
|
|
6TRT
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant S180C/T742C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TERBIUM(III) ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS8
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant G177C/A786C. | | Descriptor: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M, Chandran, A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TRF
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) purified from cells treated with kifunensine. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.106 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS2
 
 | | Truncated version of Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) lacking domain TRXL2 (417-650). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (5.74 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
4DVH
 
 | | Crystal structure of Trypanosoma cruzi mitochondrial iron superoxide dismutase | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Larrieux, N, Buschiazzo, A. | | Deposit date: | 2012-02-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Molecular Basis of the Peroxynitrite-mediated Nitration and Inactivation of Trypanosoma cruzi Iron-Superoxide Dismutases (Fe-SODs) A and B: DISPARATE SUSCEPTIBILITIES DUE TO THE REPAIR OF TYR35 RADICAL BY CYS83 IN Fe-SODB THROUGH INTRAMOLECULAR ELECTRON TRANSFER.
J.Biol.Chem., 289, 2014
|
|
2LT2
 
 | |
