4GS3
 
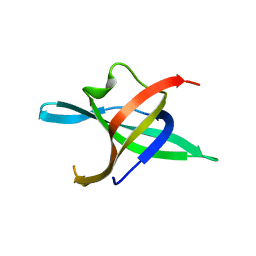 | | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tencongensis solved ab initio | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Liebschner, D, Brzezinski, K, Dauter, M, Dauter, Z, Nowak, M, Kur, J, Olszewski, M. | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tengcongensis solved ab initio.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3O4P
 
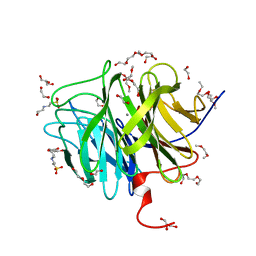 | | DFPase at 0.85 Angstrom resolution (H atoms included) | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Liebschner, D, Elias, M, Koepke, J, Lecomte, C, Guillot, B, Jelsch, C, Chabriere, E. | | Deposit date: | 2010-07-27 | | Release date: | 2011-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Hydrogen atoms in protein structures: high-resolution X-ray diffraction structure of the DFPase.
BMC Res Notes, 6, 2013
|
|
4I8K
 
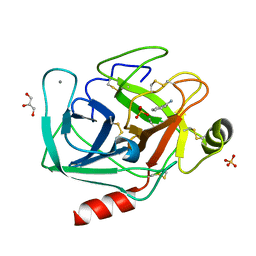 | | Bovine trypsin at 0.85 resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I8H
 
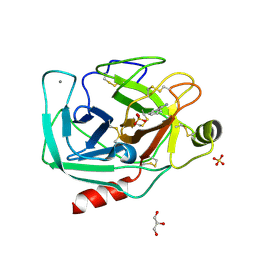 | | Bovine trypsin at 0.75 resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I8J
 
 | | Bovine trypsin at 0.87 A resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I8L
 
 | | Bovine trypsin at 0.87 resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I8G
 
 | | Bovine trypsin at 0.8 resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4F1U
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with phosphate at pH 4.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROGENPHOSPHATE ION, Putative alkaline phosphatase, ... | | Authors: | Liebschner, D, Elias, M, Tawfik, D.S, Moniot, S, Fournier, B, Scott, K, Jelsch, C, Guillot, B, Lecomte, C, Chabriere, E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
4F1V
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with phosphate at pH 8.5 | | Descriptor: | HYDROGENPHOSPHATE ION, Putative alkaline phosphatase, SULFATE ION | | Authors: | Liebschner, D, Elias, M, Tawfik, D.S, Moniot, S, Fournier, B, Scott, K, Jelsch, C, Guillot, B, Lecomte, C, Chabriere, E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZN
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - GalNAc complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Alpha-N-acetylgalactosaminidase, CALCIUM ION, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZP
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - ligand free | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | Descriptor: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5GU5
 
 | |
7LM4
 
 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8G9B
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; compressed filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8G8F
 
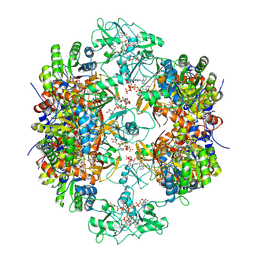 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8XWX
 
 | |
9AZB
 
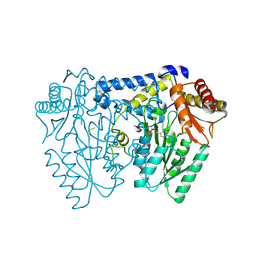 | | Crystal structure of LolTv5 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
9AZA
 
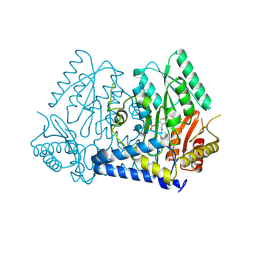 | | Crystal structure of LolTv4 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
8ERX
 
 | | Structure of chimeric HLA-A*11:01-A*02:01 bound to HIV-1 RT peptide | | Descriptor: | Beta-2-microglobulin, HIV-1 RT, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
