8SGZ
 
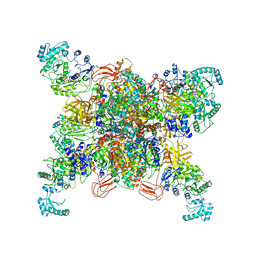 | | Leishmania tarentolae propionyl-CoA carboxylase (alpha-6-beta-6) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-coa carboxylase beta chain, putative, ... | | Authors: | Lee, J.K.J, Liu, Y.T, Hu, J.J, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2023-04-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
J Struct Biol X, 7, 2023
|
|
8SGX
 
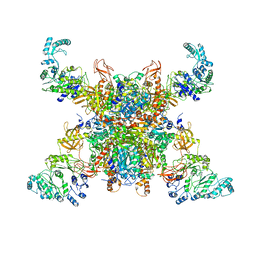 | | Leishmania tarentolae propionyl-CoA carboxylase (alpha-4-beta-6) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-coa carboxylase beta chain, putative, ... | | Authors: | Lee, J.K.J, Liu, Y.T, Hu, J.J, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2023-04-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
J Struct Biol X, 7, 2023
|
|
8SGY
 
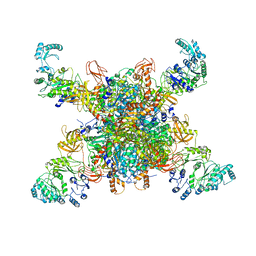 | | Leishmania tarentolae propionyl-CoA carboxylase (alpha-5-beta-6) | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-coa carboxylase beta chain, putative, ... | | Authors: | Lee, J.K.J, Liu, Y.T, Hu, J.J, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2023-04-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.62 Å) | | Cite: | CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
J Struct Biol X, 7, 2023
|
|
1JXV
 
 | | Crystal Structure of Human Nucleoside Diphosphate Kinase A | | Descriptor: | Nucleoside Diphosphate Kinase A | | Authors: | Min, K, Song, H.K, Chang, C, Kim, S.Y, Lee, K.J, Suh, S.W. | | Deposit date: | 2001-09-10 | | Release date: | 2002-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human nucleoside diphosphate kinase A, a metastasis suppressor.
Proteins, 46, 2002
|
|
1JTD
 
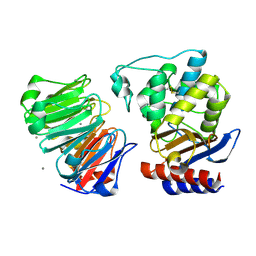 | | Crystal structure of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase | | Descriptor: | CALCIUM ION, TEM-1 beta-lactamase, beta-lactamase inhibitor protein II | | Authors: | Lim, D.C, Park, H.U, De Castro, L, Kang, S.G, Lee, H.S, Jensen, S, Lee, K.J, Strynadka, N.C.J. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
3QWZ
 
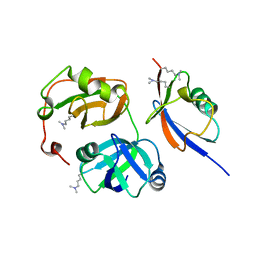 | | Crystal structure of FAF1 UBX-p97N-domain complex | | Descriptor: | FAS-associated factor 1, Transitional endoplasmic reticulum ATPase | | Authors: | Park, J.K, Jeon, H, Lee, J.J, Kim, K.H, Lee, K.J, Kim, E.E. | | Deposit date: | 2011-02-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of the interaction between FAF1 UBX and p97
To be Published
|
|
3QX1
 
 | | Crystal structure of FAF1 UBX domain | | Descriptor: | FAS-associated factor 1, SULFATE ION | | Authors: | Park, J.K, Jeon, H, Lee, J.J, Kim, K.H, Lee, K.J, Kim, E.E. | | Deposit date: | 2011-03-01 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissection of the interaction between FAF1 UBX and p97
To be Published
|
|
1P1A
 
 | | NMR structure of ubiquitin-like domain of hHR23B | | Descriptor: | UV excision repair protein RAD23 homolog B | | Authors: | Ryu, K.S, Lee, K.J, Bae, S.H, Kim, B.K, Kim, K.A, Choi, B.S. | | Deposit date: | 2003-04-11 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a
J.Biol.Chem., 278, 2003
|
|
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8F3D
 
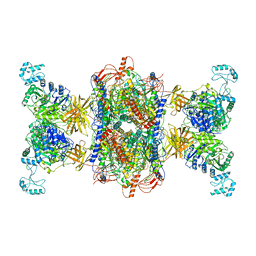 | | 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase alpha-subunit, 3-methylcrotonyl-CoA carboxylase beta-subunit, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8F41
 
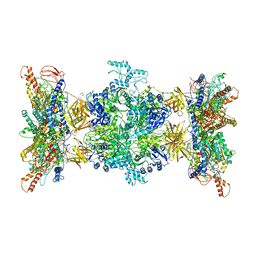 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
3MVE
 
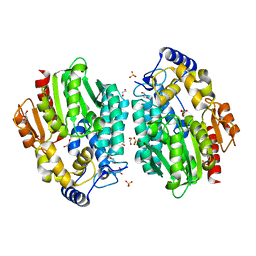 | | Crystal structure of a novel pyruvate decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UPF0255 protein VV1_0328 | | Authors: | Cha, S.S, Jeong, C.S, An, Y.J. | | Deposit date: | 2010-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | FrsA functions as a cofactor-independent decarboxylase to control metabolic flux.
Nat.Chem.Biol., 7, 2011
|
|
3OUR
 
 | |
6NDL
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
6ORU
 
 | |
4U66
 
 | |
1ZKJ
 
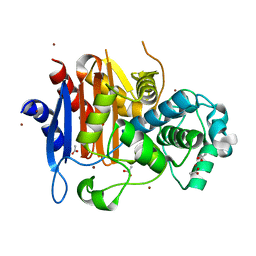 | | Structural Basis for the Extended Substrate Spectrum of CMY-10, a Plasmid-Encoded Class C beta-lactamase | | Descriptor: | ACETIC ACID, ZINC ION, extended-spectrum beta-lactamase | | Authors: | Cha, S.S, Jung, H.I, An, Y.J, Lee, S.H. | | Deposit date: | 2005-05-03 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase.
Mol.Microbiol., 60, 2006
|
|
1JTG
 
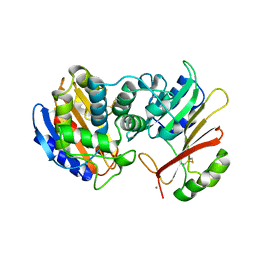 | | CRYSTAL STRUCTURE OF TEM-1 BETA-LACTAMASE / BETA-LACTAMASE INHIBITOR PROTEIN COMPLEX | | Descriptor: | BETA-LACTAMASE INHIBITORY PROTEIN, BETA-LACTAMASE TEM, CALCIUM ION | | Authors: | Strynadka, N.C.J, Jensen, S.E, Alzari, P.M, James, M.N. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
7FGN
 
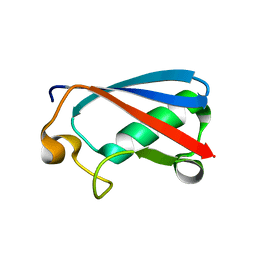 | | The crystal structure of the FAF1 UBL1 | | Descriptor: | FAS-associated factor 1 | | Authors: | Kim, E.E, ParK, J.K, Shin, S.C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | The complex of Fas-associated factor 1 with Hsp70 stabilizes the adherens junction integrity by suppressing RhoA activation
J Mol Cell Biol, 14, 2022
|
|
7FGM
 
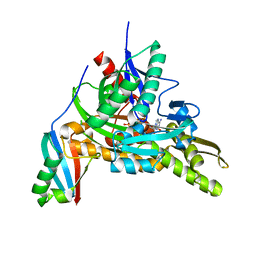 | | The complex crystals structure of the FAF1 UBL1_L-Hsp70 NBD with ADP and phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAS-associated factor 1, Heat shock 70 kDa protein 1A, ... | | Authors: | Kim, E.E, ParK, J.K, Shin, S.C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The complex of Fas-associated factor 1 with Hsp70 stabilizes the adherens junction integrity by suppressing RhoA activation
J Mol Cell Biol, 14, 2022
|
|
