6AH3
 
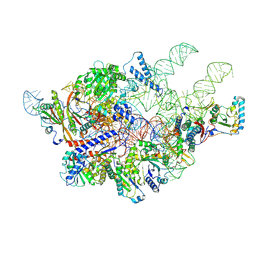 | | Cryo-EM structure of yeast Ribonuclease P with pre-tRNA substrate | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
6AGB
 
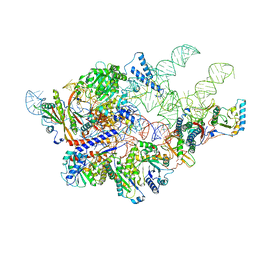 | | Cryo-EM structure of yeast Ribonuclease P | | Descriptor: | RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, Ribonuclease P protein subunit RPR2, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
7C7A
 
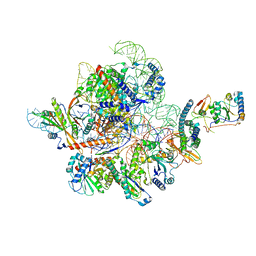 | | Cryo-EM structure of yeast Ribonuclease MRP with substrate ITS1 | | Descriptor: | Internal Transcribed Spacer 1, MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
7C79
 
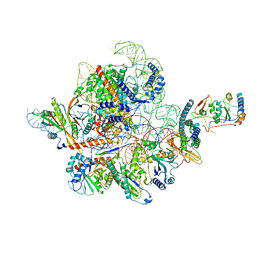 | | Cryo-EM structure of yeast Ribonuclease MRP | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease MRP RNA subunit NME1, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
2F9Q
 
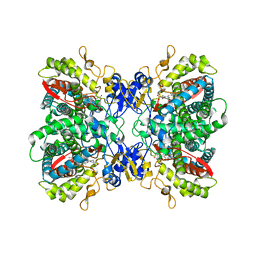 | | Crystal Structure of Human Cytochrome P450 2D6 | | Descriptor: | Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Rowland, P. | | Deposit date: | 2005-12-06 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Crystal Structure of Human Cytochrome P450 2D6
J.Biol.Chem., 281, 2006
|
|
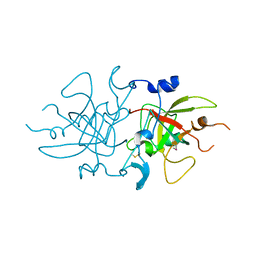
8A4O
 
 | |
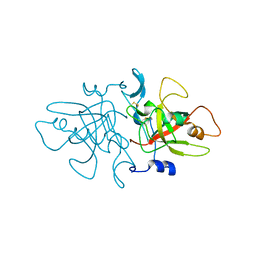
8A14
 
 | |
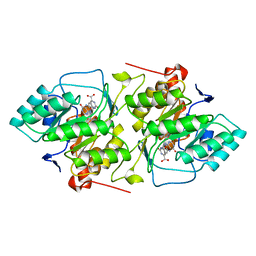
2DOR
 
 | |
1EP3
 
 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B. DATA COLLECTED UNDER CRYOGENIC CONDITIONS. | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
1DOR
 
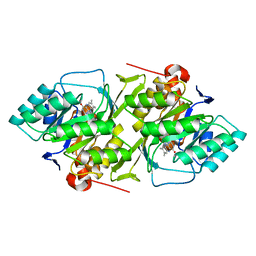 | | DIHYDROOROTATE DEHYDROGENASE A FROM LACTOCOCCUS LACTIS | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE | | Authors: | Rowland, P, Larsen, S. | | Deposit date: | 1997-01-14 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the flavin containing enzyme dihydroorotate dehydrogenase A from Lactococcus lactis.
Structure, 5, 1997
|
|
1EP2
 
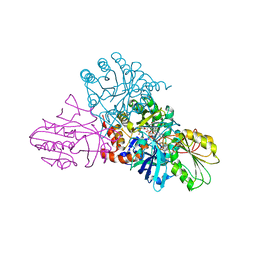 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B COMPLEXED WITH OROTATE | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
1EP1
 
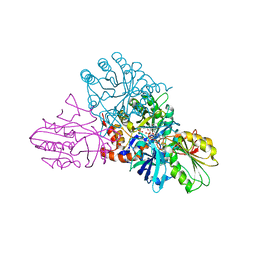 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
4GE3
 
 | | Schizosaccharomyces pombe DJ-1 T114V mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein C22E12.03c | | Authors: | Madzelan, P, Labunska, T, Wilson, M.A. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influence of peptide dipoles and hydrogen bonds on reactive cysteine pK(a) values in fission yeast DJ-1.
Febs J., 279, 2012
|
|
4GE0
 
 | | Schizosaccharomyces pombe DJ-1 T114P mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein C22E12.03c | | Authors: | Madzelan, P, Labunska, T, Wilson, M.A. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of peptide dipoles and hydrogen bonds on reactive cysteine pK(a) values in fission yeast DJ-1.
Febs J., 279, 2012
|
|
7AX4
 
 | |
7Q7K
 
 | | JAK2 in complex with 4-(2-amino-8-methoxyquinazolin-6-yl)phenol | | Descriptor: | 4-(2-azanyl-8-methoxy-quinazolin-6-yl)phenol, Tyrosine-protein kinase JAK2 | | Authors: | Rowland, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Investigation of Janus Kinase (JAK) Inhibitors for Lung Delivery and the Importance of Aldehyde Oxidase Metabolism.
J.Med.Chem., 65, 2022
|
|
7Q7L
 
 | |
7Q7I
 
 | |
7Q7W
 
 | |
8OLU
 
 | |
7POT
 
 | | PI3 kinase delta in complex with N-[5-(3,6-dihydro-2H-pyran-4-yl)-2-methoxypyridin-3-yl]benzenesulfonamide | | Descriptor: | N-[5-(3,6-dihydro-2H-pyran-4-yl)-2-methoxy-pyridin-3-yl]benzenesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Rowland, P, Convery, M. | | Deposit date: | 2021-09-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Discovery of GSK251: A Highly Potent, Highly Selective, Orally Bioavailable Inhibitor of PI3K delta with a Novel Binding Mode.
J.Med.Chem., 64, 2021
|
|
7POR
 
 | |
7POS
 
 | | PI3 kinase delta in complex with 5-(3,6-dihydro-2H-pyran-4-yl)-2-methoxy-N-(5-{3-[4-(propan-2-yl)piperazin-1-yl]prop-1-yn-1-yl}pyridin-3-yl)pyridine-3-sulfonamide | | Descriptor: | 5-(3,6-dihydro-2~{H}-pyran-4-yl)-2-methoxy-~{N}-[5-[3-(4-propan-2-ylpiperazin-1-yl)prop-1-ynyl]pyridin-3-yl]pyridine-3-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Rowland, P, Convery, M. | | Deposit date: | 2021-09-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Discovery of GSK251: A Highly Potent, Highly Selective, Orally Bioavailable Inhibitor of PI3K delta with a Novel Binding Mode.
J.Med.Chem., 64, 2021
|
|
7POP
 
 | | PI3 kinase delta in complex with 5-[3,6-dihydro-2H-pyran-4-yl]-2-methoxy-N-[2-methylpyridin-4-yl]pyridine-3-sulfonamide | | Descriptor: | 5-[3,6-dihydro-2H-pyran-4-yl]-2-methoxy-N-[2-methylpyridin-4-yl]pyridine-3-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Rowland, P, Convery, M. | | Deposit date: | 2021-09-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Discovery of GSK251: A Highly Potent, Highly Selective, Orally Bioavailable Inhibitor of PI3K delta with a Novel Binding Mode.
J.Med.Chem., 64, 2021
|
|
2O0U
 
 | | Crystal structure of human JNK3 complexed with N-{3-cyano-6-[3-(1-piperidinyl)propanoyl]-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl}-1-naphthalenecarboxamide | | Descriptor: | Mitogen-activated protein kinase 10, N-{3-CYANO-6-[3-(1-PIPERIDINYL)PROPANOYL]-4,5,6,7-TETRAHYDROTHIENO[2,3-C]PYRIDIN-2-YL}1-NAPHTHALENECARBOXAMIDE | | Authors: | Rowland, P, Somers, D. | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N-(3-Cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)amides as potent, selective, inhibitors of JNK2 and JNK3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
