1AUZ
 
 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BUZ
 
 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1M7L
 
 | | Solution Structure of the Coiled-Coil Trimerization Domain from Lung Surfactant Protein D | | Descriptor: | Pulmonary surfactant-associated protein D | | Authors: | Kovacs, H, O'Donoghue, S.I, Hoppe, H.-J, Comfort, D, Reid, K.B.M, Campbell, I.D, Nilges, M. | | Deposit date: | 2002-07-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the coiled-coil trimerization domain from lung surfactant protein D
J.BIOMOL.NMR, 24, 2002
|
|
1GDC
 
 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
2W0D
 
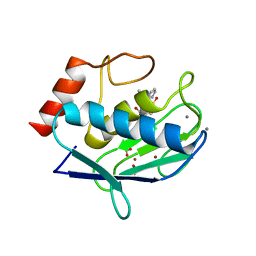 | | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? A Case Study of Metalloproteinases. | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Isaksson, J, Nystrom, S, Derbyshire, D.J, Wallberg, H, Agback, T, Kovacs, H, Bertini, I, Felli, I.C. | | Deposit date: | 2008-08-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Does a Fast Nuclear Magnetic Resonance Spectroscopy- and X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information of Ligand-Protein Complexes? a Case Study of Metalloproteinases.
J.Med.Chem., 52, 2009
|
|
2GDA
 
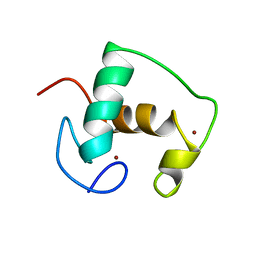 | | REFINED SOLUTION STRUCTURE OF THE GLUCOCORTICOID RECEPTOR DNA-BINDING DOMAIN | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Baumann, H, Paulsen, K, Kovacs, H, Berglund, H, Wright, A.P.H, Gustafsson, J.-A, Hard, T. | | Deposit date: | 1994-03-15 | | Release date: | 1994-06-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the glucocorticoid receptor DNA-binding domain.
Biochemistry, 32, 1993
|
|
5QTN
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with SS-4432 | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoro-1H-pyrrolo[2,3-b]pyridine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QTQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with FS-1169 | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoroquinazolin-4(3H)-one, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QTO
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with 1R-0641 | | Descriptor: | 1,2-ETHANEDIOL, 3-(difluoromethyl)-8-(trifluoromethyl)[1,2,4]triazolo[4,3-a]pyridine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QTL
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with GB-0804 | | Descriptor: | 1,2-ETHANEDIOL, 4-(trifluoromethyl)pyrimidin-2-amine, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-10-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QTP
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with AE-0227 | | Descriptor: | 1,2-ETHANEDIOL, 8-chloro-6-(trifluoromethyl)imidazo[1,2-a]pyridine-7-carbonitrile, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QTR
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with FS-3764 | | Descriptor: | 1,2-ETHANEDIOL, 6-fluoroquinazolin-4(3H)-one, MAGNESIUM ION, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-10-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QTM
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with FS-2639 | | Descriptor: | 1,2-ETHANEDIOL, 7-fluoroquinazolin-4(3H)-one, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QTS
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with 8J-537S | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2-(methylsulfanyl)-6-(trifluoromethyl)pyrimidin-4(3H)-one, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Krojer, T, Kovacs, H, Moriaud, F, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2019-08-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1RO3
 
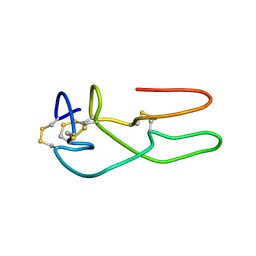 | | New structural insights on short disintegrin echistatin by NMR | | Descriptor: | Disintegrin echistatin | | Authors: | Monleon, D, Esteve, V, Calvete, J.J, Marcinkiewicz, C, Celda, B. | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Conformation and concerted dynamics of the integrin-binding site and the C-terminal region of echistatin revealed by homonuclear NMR
Biochem.J., 387, 2005
|
|
1ZBJ
 
 | |
7PDU
 
 | |
6H54
 
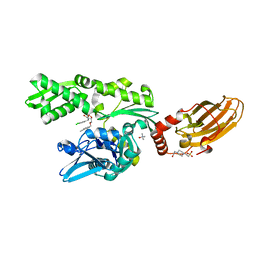 | | CRYSTAL STRUCTURE OF BOVINE HSC70(AA1-554)E213A/D214A IN COMPLEX WITH INHIBITOR VER155008 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, GLYCEROL, ... | | Authors: | Plank, C, Zehe, M, Grimm, C, Sotriffer, C. | | Deposit date: | 2018-07-23 | | Release date: | 2019-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Combined In-Solution Fragment Screening and Crystallographic Binding-Mode Analysis with a Two-Domain Hsp70 Construct.
Acs Chem.Biol., 2024
|
|
7PLK
 
 | |
7O6R
 
 | |
7ODI
 
 | |
7ODD
 
 | |
7ODB
 
 | | Crystal structure of bovine Hsc70(aa1-554)E213A/D214A in complex with triazine-derivative | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-methyl-5-sulfanylidene-2H-1,2,4-triazin-3-one, GLYCEROL, ... | | Authors: | Zehe, M, Grimm, C, Sotriffer, C. | | Deposit date: | 2021-04-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Combined In-Solution Fragment Screening and Crystallographic Binding-Mode Analysis with a Two-Domain Hsp70 Construct.
Acs Chem.Biol., 2024
|
|
