1IOZ
 
 | | Crystal Structure of the C-HA-RAS Protein Prepared by the Cell-Free Synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Kigawa, T, Yamaguchi-Nunokawa, E, Kodama, K, Matsuda, T, Yabuki, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selenomethionine incorporation into a protein by cell-free synthesis
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
3UFZ
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the universal genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
8KCQ
 
 | | Solution structures of the N-terminal divergent caplonin homology (NN-CH) domains of human intraflagellar transport protein 54 | | Descriptor: | TRAF3-interacting protein 1 | | Authors: | Dang, W, Kuwasako, K, He, F, Takahashi, M, Tsuda, K, Nagata, T, Tanaka, A, Kobayashi, N, Kigawa, T, Guentert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2023-08-08 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structure of the N-terminal divergent calponin homology (NN-CH) domain of human intraflagellar transport protein 54.
Biomol.Nmr Assign., 18, 2024
|
|
5H0Q
 
 | | Crystal structure of lipid binding protein Nakanori at 1.5A | | Descriptor: | Lipid binding protein | | Authors: | Makino, A, Abe, M, Ishitsuka, R, Murate, M, Kishimoto, T, Sakai, S, Hullin-Matsuda, F, Shimada, Y, Inaba, T, Miyatake, H, Tanaka, H, Kurahashi, A, Pack, C.G, Kasai, R.S, Kubo, S, Schieber, N.L, Dohmae, N, Tochio, N, Hagiwara, K, Sasaki, Y, Aida, Y, Fujimori, F, Kigawa, T, Nishikori, K, Parton, R.G, Kusumi, A, Sako, Y, Anderluh, G, Yamashita, M, Kobayashi, T, Greimel, P, Kobayashi, T. | | Deposit date: | 2016-10-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C
FASEB J., 31, 2017
|
|
3UG0
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the simplified genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
1BW6
 
 | | HUMAN CENTROMERE PROTEIN B (CENP-B) DNA BINDIGN DOMAIN RP1 | | Descriptor: | PROTEIN (CENTROMERE PROTEIN B) | | Authors: | Iwahara, J, Kigawa, T, Kitagawa, K, Masumoto, H, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-09-30 | | Release date: | 1998-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A helix-turn-helix structure unit in human centromere protein B (CENP-B).
EMBO J., 17, 1998
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
5B1O
 
 | | DHp domain structure of EnvZ P248A mutant | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
5B1N
 
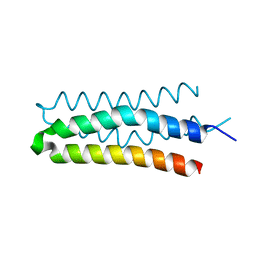 | | DHp domain structure of EnvZ from Escherichia coli | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
1RRB
 
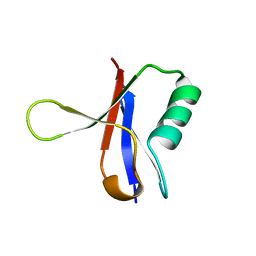 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
1N27
 
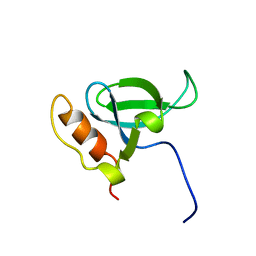 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | Descriptor: | Hepatoma-derived growth factor, related protein 3 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
7EN4
 
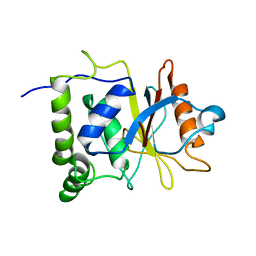 | | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase YUH1 | | Authors: | Okada, M, Tateishi, Y, Nojiri, E, Mikawa, T, Rajesh, S, Ogasawa, H, Ueda, T, Yagi, H, Kohno, T, Kigawa, T, Shimada, I, Guentert, P, Yutaka, I, Ikeya, T. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase.
To Be Published
|
|
1R79
 
 | | Solution Structure of The C1 Domain of The Human Diacylglycerol Kinase Delta | | Descriptor: | Diacylglycerol kinase, delta, ZINC ION | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-21 | | Release date: | 2004-04-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The C1 Domain of The Human Diacylglycerol Kinase Delta
To be Published
|
|
1IXD
 
 | | Solution structure of the CAP-GLY domain from human cylindromatosis tomour-suppressor CYLD | | Descriptor: | Cylindromatosis tumour-suppressor CYLD | | Authors: | Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-06-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma
STRUCTURE, 12, 2004
|
|
1IVZ
 
 | | Solution structure of the SEA domain from murine hypothetical protein homologous to human mucin 16 | | Descriptor: | hypothetical protein 1110008I14RIK | | Authors: | Maeda, T, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-02 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16)
J.Biol.Chem., 279, 2004
|
|
2KUQ
 
 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | Fibroblast growth factor receptor substrate 3,LINKER,ALK tyrosine kinase receptor | | Authors: | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2KUP
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | 19-residue peptide from ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
7CK5
 
 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
2LC2
 
 | | Solution structure of the RXLR effector P. capsici AVR3a4 | | Descriptor: | AVR3a4 | | Authors: | Li, H, Koshiba, S, Yaeno, T, Sato, M, Watanabe, S, Harada, T, Shirasu, K, Kigawa, T. | | Deposit date: | 2011-04-12 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A PIP-binding interface in the oomycete RXLR effector AVR3A is required for its accumulation in host cells to modulate plant immunity
To be Published
|
|
1IUF
 
 | | LOW RESOLUTION SOLUTION STRUCTURE OF THE TWO DNA-BINDING DOMAINS IN Schizosaccharomyces pombe ABP1 PROTEIN | | Descriptor: | centromere abp1 protein | | Authors: | Kikuchi, J, Iwahara, J, Kigawa, T, Murakami, Y, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-04 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure determination of the two DNA-binding domains in the Schizosaccharomyces pombe Abp1 protein by a combination of dipolar coupling and diffusion anisotropy restraints.
J.Biomol.NMR, 22, 2002
|
|
1J03
 
 | | Solution structure of a putative steroid-binding protein from Arabidopsis | | Descriptor: | putative steroid binding protein | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Terada, T, Shirouzu, M, Seki, M, Shinozaki, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-29 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis homologue of the mammalian membrane-associated progesterone receptor
To be Published
|
|
1PMS
 
 | | PLECKSTRIN HOMOLOGY DOMAIN OF SON OF SEVENLESS 1 (SOS1) WITH GLYCINE-SERINE ADDED TO THE N-TERMINUS, NMR, 20 STRUCTURES | | Descriptor: | SOS 1 | | Authors: | Koshiba, S, Kigawa, T, Kim, J, Shirouzu, M, Bowtell, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-02-18 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the pleckstrin homology domain of mouse Son-of-sevenless 1 (mSos1).
J.Mol.Biol., 269, 1997
|
|
1KOY
 
 | | NMR structure of DFF-C domain | | Descriptor: | DNA fragmentation factor alpha subunit | | Authors: | Fukushima, K, Kikuchi, J, Koshiba, S, Kigawa, T, Kuroda, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-25 | | Release date: | 2002-09-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DFF-C domain of DFF45/ICAD. A structural basis for the regulation of apoptotic DNA fragmentation.
J.Mol.Biol., 321, 2002
|
|
