8HVP
 
 | | STRUCTURE AT 2.5-ANGSTROMS RESOLUTION OF CHEMICALLY SYNTHESIZED HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE COMPLEXED WITH A HYDROXYETHYLENE*-BASED INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR VAL-SER-GLN-ASN-LEU-PSI(CH(OH)-CH2)-VAL-ILE-VAL (U-85548E) | | Authors: | Jaskolski, M, Miller, M, Tomasselli, A.G, Sawyer, T.K, Staples, D.G, Heinrikson, R.L, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-10-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor.
Biochemistry, 30, 1991
|
|
1HO3
 
 | | CRYSTAL STRUCTURE ANALYSIS OF E. COLI L-ASPARAGINASE II (Y25F MUTANT) | | Descriptor: | ASPARTIC ACID, L-ASPARAGINASE II | | Authors: | Jaskolski, M, Kozak, M, Lubkowski, P, Palm, J.G, Wlodawer, A. | | Deposit date: | 2000-12-08 | | Release date: | 2001-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two highly homologous bacterial L-asparaginases: a case of enantiomorphic space groups.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HFK
 
 | | Asparaginase from Erwinia chrysanthemi, hexagonal form with weak sulfate | | Descriptor: | L-ASPARAGINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Lubkowski, J, Palm, G.J, Kozak, M, Jaskolski, M, Wlodawer, A. | | Deposit date: | 2000-12-05 | | Release date: | 2000-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of Two Highly Homologous Bacterial L-Asparaginases: A Case of Enantiomorphic Space Groups
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HFJ
 
 | | Asparaginase from Erwinia chrysanthemi, hexagonal form with sulfate | | Descriptor: | L-ASPARAGINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Palm, G.J, Lubkowski, J, Kozak, M, Jaskolski, M, Wlodawer, A. | | Deposit date: | 2000-12-05 | | Release date: | 2000-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Two Highly Homologous Bacterial L-Asparaginases: A Case of Enantiomorphic Space Groups
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3SQF
 
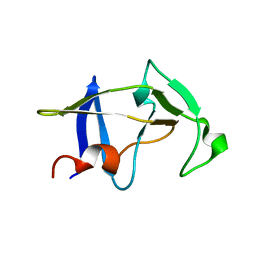 | | Crystal structure of monomeric M-PMV retroviral protease | | Descriptor: | Protease | | Authors: | Jaskolski, M, Kazmierczyk, M, Gilski, M, Krzywda, S, Pichova, I, Zabranska, H, Khatib, F, DiMaio, F, Cooper, S, Thompson, J, Popovic, Z, Baker, D, Group, Foldit Contenders | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6324 Å) | | Cite: | Crystal structure of a monomeric retroviral protease solved by protein folding game players.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2RSP
 
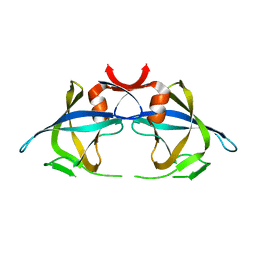 | |
5I8F
 
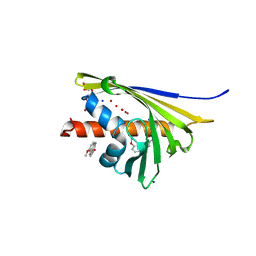 | | Crystal structure of St. John's wort Hyp-1 protein in complex with melatonin | | Descriptor: | GLYCEROL, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Jaskolski, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin.
Front Plant Sci, 7, 2016
|
|
1R4C
 
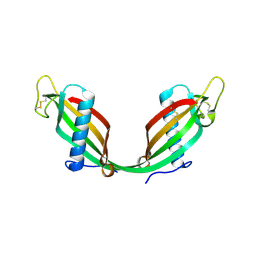 | |
3IE5
 
 | | Crystal structure of Hyp-1 protein from Hypericum perforatum (St John's wort) involved in hypericin biosynthesis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Michalska, K, Fernandes, H, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin
J.Struct.Biol., 169, 2010
|
|
4R15
 
 | | High-resolution crystal structure of Z-DNA in complex with Cr3+ cations | | Descriptor: | CHROMIUM ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2014-08-04 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High-resolution crystal structure of Z-DNA in complex with Cr(3+) cations.
J.Biol.Inorg.Chem., 20, 2015
|
|
2ZAK
 
 | | Orthorhombic crystal structure of precursor E. coli isoaspartyl peptidase/L-asparaginase (EcAIII) with active-site T179A mutation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, L-asparaginase precursor, ... | | Authors: | Michalska, K, Hernandez-Santoyo, A, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal packing of plant-type L-asparaginase from Escherichia coli
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6S1U
 
 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1W
 
 | | Crystal structure of dimeric M-PMV protease D26N mutant | | Descriptor: | Gag-Pro-Pol polyprotein | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7NEN
 
 | |
6S1V
 
 | | Crystal structure of dimeric M-PMV protease D26N mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5MXB
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin | | Descriptor: | Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, SODIUM ION, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
5MXW
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin and trans-zeatin. | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
2QIM
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, CALCIUM ION, GLYCEROL, ... | | Authors: | Fernandes, H.C, Pasternak, O, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2007-07-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin.
J.Mol.Biol., 378, 2008
|
|
5JZQ
 
 | |
7BGU
 
 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, peptidomimetic inhibitor | | Authors: | Wosicki, S, Gilski, M, Kazmierczyk, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
7BGT
 
 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | ACETATE ION, Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, ... | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
4PV2
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ and Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, NITRATE ION, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Clavel, D, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PU6
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ cations | | Descriptor: | ASPARTIC ACID, L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PV3
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, SODIUM ION | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8RUG
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV C189A mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
