8R2C
 
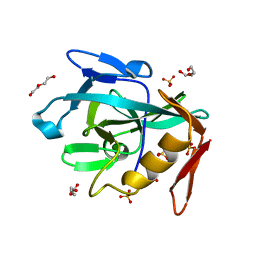 | | Crystal structure of the Vint domain from Tetrahymena thermophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, von willebrand factor type A (VWA) domain was originally protein | | Authors: | Iwai, H, Beyer, H.M, Johannson, J.E, Li, M, Wlodawer, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of the Vint domain from Tetrahymena thermophila suggests a ligand-regulated cleavage mechanism by the HINT fold.
Febs Lett., 598, 2024
|
|
1VD0
 
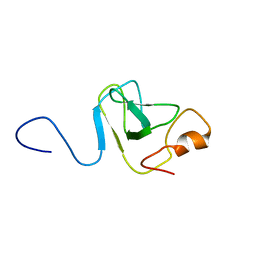 | | Capsid stabilizing protein GPD, NMR, 20 Structures | | Descriptor: | Head decoration protein | | Authors: | Iwai, H, Forrer, P, Pluckthun, A, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-17 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the monomeric form of the bacteriophage lambda capsid stabilizing protein gpD.
J.Biomol.Nmr, 31, 2005
|
|
1SQ8
 
 | |
8RD6
 
 | |
5O9I
 
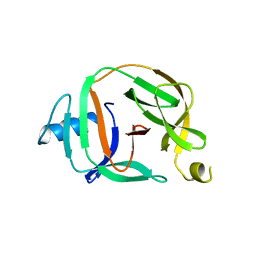 | | Crystal structure of transcription factor IIB Mvu mini-intein | | Descriptor: | Transcription initiation factor IIB,Transcription initiation factor IIB | | Authors: | Mikula, K.M, Iwai, H, Li, M, Wlodawer, A. | | Deposit date: | 2017-06-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Persistence of Homing Endonucleases in Transcription Factor IIB Inteins.
J. Mol. Biol., 429, 2017
|
|
5O9J
 
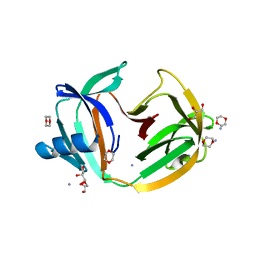 | | Crystal structure of transcription factor IIB Mja mini-intein | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMMONIUM ION, ... | | Authors: | Mikula, K.M, Iwai, H, Zhou, D, Wlodawer, A. | | Deposit date: | 2017-06-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Persistence of Homing Endonucleases in Transcription Factor IIB Inteins.
J. Mol. Biol., 429, 2017
|
|
7OEC
 
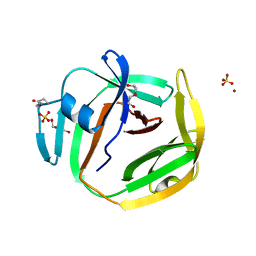 | | Crystal structure of an intein from a hyperthermophile | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA polymerase II large subunit, SULFATE ION, ... | | Authors: | Hannes, B, Hiltunen, M, Iwai, H. | | Deposit date: | 2021-05-03 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mini-Intein Structures from Extremophiles Suggest a Strategy for Finding Novel Robust Inteins.
Microorganisms, 9, 2021
|
|
5OBN
 
 | | NMR solution structure of U11/U12 65K protein's C-terminal RRM domain (381-516) | | Descriptor: | RNA-binding protein 40 | | Authors: | Norppa, A.J, Kauppala, T.M, Heikkinen, H.A, Verma, B, Iwai, H, Frilander, M.J. | | Deposit date: | 2017-06-28 | | Release date: | 2018-01-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Mutations in the U11/U12-65K protein associated with isolated growth hormone deficiency lead to structural destabilization and impaired binding of U12 snRNA.
RNA, 24, 2018
|
|
7QST
 
 | |
7QSU
 
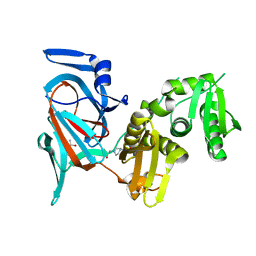 | |
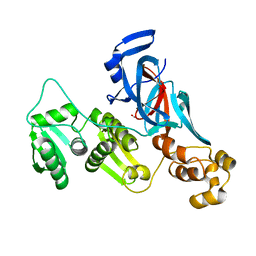
7QSS
 
 | |
7QIL
 
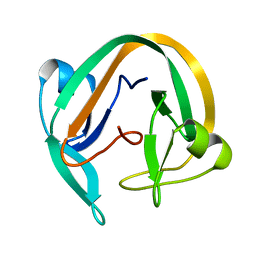 | |
8BFG
 
 | | Solution structure of human apo/Calmodulin G113R (G114R) | | Descriptor: | Calmodulin-1 | | Authors: | Wimmer, R, Holler, C.V, Petersson, N.M, Brohus, M.B, Niemelae, M, Overgaard, M.T, Iwai, H. | | Deposit date: | 2022-10-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Allosteric changes in protein stability and dynamics as pathogenic mechanism for calmodulin variants not affecting Ca 2+ coordinating residues.
Cell Calcium, 117, 2023
|
|
8BD2
 
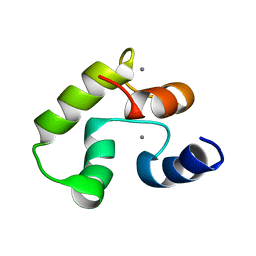 | | Calcium-bound Calmodulin variant G113R | | Descriptor: | CALCIUM ION, Calmodulin-3 | | Authors: | Wimmer, R, Holler, C.V, Petersson, N.M, Iwai, H, Niemelae, M.A, Brohus, M, Overgaard, M.T. | | Deposit date: | 2022-10-18 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Allosteric changes in protein stability and dynamics as pathogenic mechanism for calmodulin variants not affecting Ca 2+ coordinating residues.
Cell Calcium, 117, 2023
|
|
1KP5
 
 | | Cyclic Green Fluorescent Protein | | Descriptor: | Green Fluorescent Protein, SULFATE ION | | Authors: | Hofmann, A, Iwai, H, Plueckthun, A, Wlodawer, A. | | Deposit date: | 2001-12-28 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cyclized green fluorescent protein.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6FFQ
 
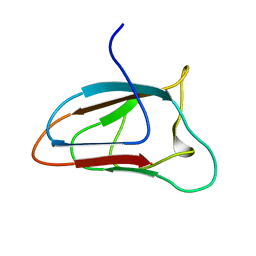 | | Solution NMR structure of CBM64 from S.thermophila | | Descriptor: | Glycosyl hydrolase family 5 cellulase CBM64 | | Authors: | Heikkinen, H.A, Iwai, H. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Determinations of Small Proteins Using only One Fractionally 20% 13 C- and Uniformly 100% 15 N-Labeled Sample.
Molecules, 26, 2021
|
|
6FFU
 
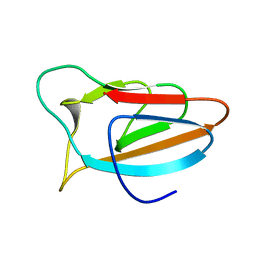 | |
6FIP
 
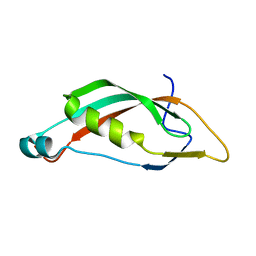 | |
1OAW
 
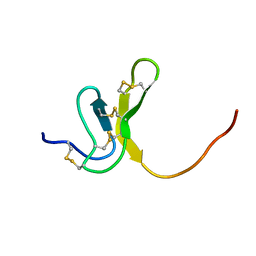 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
1OAV
 
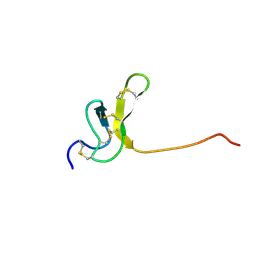 | | OMEGA-AGATOXIN IVA | | Descriptor: | OMEGA-AGATOXIN IVA | | Authors: | Kim, J.I, Konishi, S, Iwai, H, Kohno, T, Gouda, H, Shimada, I, Sato, K, Arata, Y. | | Deposit date: | 1995-06-28 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium channel antagonist omega-agatoxin IVA: consensus molecular folding of calcium channel blockers.
J.Mol.Biol., 250, 1995
|
|
5N2N
 
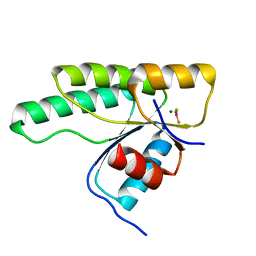 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana complexed with Mg2+ and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Histidine kinase CKI1, MAGNESIUM ION | | Authors: | Otrusinova, O, Demo, G, Padrta, P, Jasenakova, Z, Pekarova, B, Gelova, Z, Szmitkowska, A, Kaderavek, P, Jansen, S, Zachrdla, M, Klumpler, T, Marek, J, Hritz, J, Janda, L, Iwai, H, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2017-02-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
4KL6
 
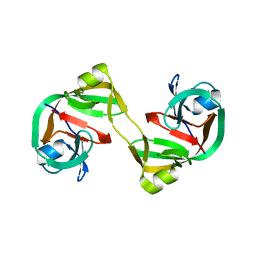 | | Crystal structure of dimeric form of NpuDnaE intein | | Descriptor: | DNA-directed DNA polymerase,Nucleic acid binding, OB-fold, tRNA/helicase-type | | Authors: | Aranko, A.S, Oeemig, J.S, Kajander, T, Iwai, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intermolecular domain swapping induces intein-mediated protein alternative splicing.
Nat.Chem.Biol., 9, 2013
|
|
5LW8
 
 | |
4KL5
 
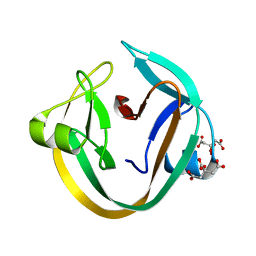 | | Crystal structure of NpuDnaE intein | | Descriptor: | CITRIC ACID, DNA polymerase III, alpha subunit, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Kajander, T, Iwai, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Intermolecular domain swapping induces intein-mediated protein alternative splicing.
Nat.Chem.Biol., 9, 2013
|
|
6SLY
 
 | |
