4QQB
 
 | | Structural basis for the assembly of the SXL-UNR translation regulatory complex | | Descriptor: | Protein sex-lethal, Upstream of N-ras, isoform A, ... | | Authors: | Hennig, J, Popowicz, G.M, Sattler, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the assembly of the Sxl-Unr translation regulatory complex.
Nature, 515, 2014
|
|
7P4N
 
 | |
2MKL
 
 | |
2N88
 
 | | Chromodomain 3 (CD3) of cpSRP43 | | Descriptor: | Signal recognition particle 43 kDa protein, chloroplastic | | Authors: | Hennig, J, Sattler, M. | | Deposit date: | 2015-10-06 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
7ZHR
 
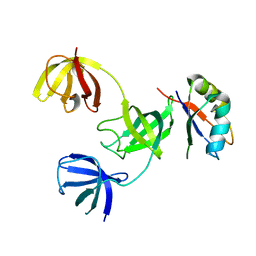 | |
7ZHH
 
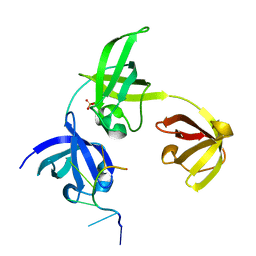 | | Complex structure of drosophila Unr CSD789 and a poly(A) RNA sequence | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, Upstream of N-ras, ... | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res., 51, 2023
|
|
8B9J
 
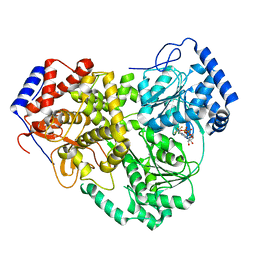 | | Cryo-EM structure of MLE in complex with ADP:AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, MAGNESIUM ION, ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
8B9L
 
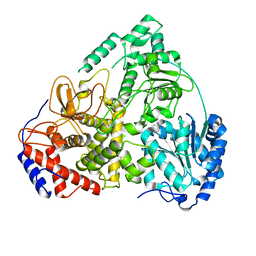 | | Cryo-EM structure of MLE | | Descriptor: | Dosage compensation regulator | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
8B9K
 
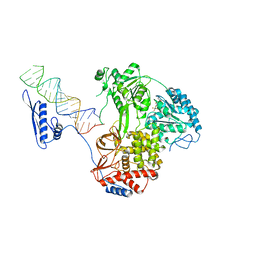 | |
8B9I
 
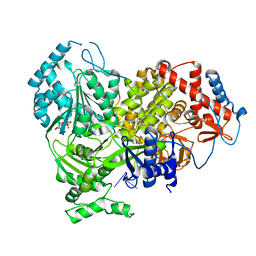 | |
8B9G
 
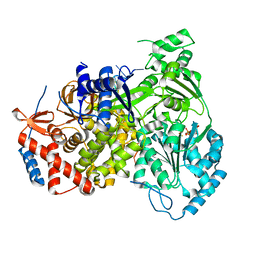 | | Cryo-EM structure of MLE in complex with ADP:AlF4 and U10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
4Q97
 
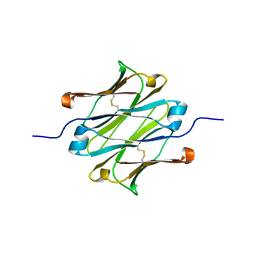 | | IgNAR antibody domain C1 | | Descriptor: | Novel antigen receptor | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q9B
 
 | | IgNAR antibody domain C2 | | Descriptor: | Novel antigen receptor | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q9C
 
 | | IgNAR antibody domain C3 | | Descriptor: | CHLORIDE ION, Novel antigen receptor, SODIUM ION, ... | | Authors: | Feige, J.M, Graewert, M.A, Marcinowski, M, Hennig, J, Behnke, J, Auslaender, D, Herold, E.M, Peschek, J, Castro, C.D, Flajnik, M.F, Hendershot, L.M, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8AMS
 
 | | Complex of human TRIM2 RING domain, UBCH5C, and Ubiquitin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Perez-Borrajero, C, Kotova, I, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
7QI0
 
 | | Crystal structure of KLK6 in complex with compound DKFZ918 | | Descriptor: | (5~{R})-3-(6-carbamimidoylpyridin-3-yl)-~{N}-[(1~{S})-1-naphthalen-1-ylpropyl]-2-oxidanylidene-1,3-oxazolidine-5-carboxamide, Kallikrein-6 | | Authors: | Jagtap, P.K.A, Baumann, A, Lohbeck, J, Isak, D, Miller, A, Hennig, J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Scalable synthesis and structural characterization of reversible KLK6 inhibitors.
Rsc Adv, 12, 2022
|
|
7QHZ
 
 | | Crystal structure of KLK6 in complex with compound DKFZ917 | | Descriptor: | (5~{R})-3-(4-carbamimidoylphenyl)-~{N}-[(1~{S})-1-naphthalen-1-ylpropyl]-2-oxidanylidene-1,3-oxazolidine-5-carboxamide, GLYCEROL, Kallikrein-6 | | Authors: | Jagtap, P.K.A, Baumann, A, Lohbeck, J, Isak, D, Miller, A, Hennig, J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Scalable synthesis and structural characterization of reversible KLK6 inhibitors.
Rsc Adv, 12, 2022
|
|
8A38
 
 | |
3UR8
 
 | | Lower-density crystal structure of potato endo-1,3-beta-glucanase | | Descriptor: | Glucan endo-1,3-beta-D-glucosidase | | Authors: | Wojtkowiak, A, Witek, K, Hennig, J, Jaskolski, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Two high-resolution structures of potato endo-1,3-beta-glucanase reveal subdomain flexibility with implications for substrate binding
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8AMR
 
 | | Coiled-coil domain of human TRIM3 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Tripartite motif-containing protein 3 | | Authors: | Perez-Borrajero, C, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
3UR7
 
 | | Higher-density crystal structure of potato endo-1,3-beta-glucanase | | Descriptor: | Glucan endo-1,3-beta-D-glucosidase, SODIUM ION | | Authors: | Wojtkowiak, A, Witek, K, Hennig, J, Jaskolski, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two high-resolution structures of potato endo-1,3-beta-glucanase reveal subdomain flexibility with implications for substrate binding
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7QFT
 
 | | Crystal structure of KLK6 in complex with compound 16a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Cha-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
7QFV
 
 | | Crystal structure of KLK6 in complex with compound 17a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Ser(Z)-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
7B96
 
 | |
8P66
 
 | |
