2TBV
 
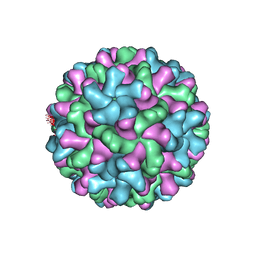 | |
3N0A
 
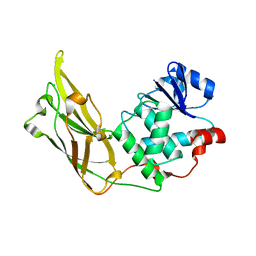 | | Crystal structure of auxilin (40-400) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein phosphatase auxilin | | Authors: | Harrison, S.C, Guan, R, Dai, H, Kirchhausen, T. | | Deposit date: | 2010-05-13 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the PTEN-like Region of Auxilin, a Detector of Clathrin-Coated Vesicle Budding.
Structure, 18, 2010
|
|
1KRI
 
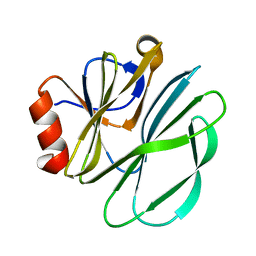 | |
4V7Q
 
 | | Atomic model of an infectious rotavirus particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Core scaffold protein, ... | | Authors: | Settembre, E.C, Chen, J.Z, Dormitzer, P.R, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2010-05-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic model of an infectious rotavirus particle.
Embo J., 30, 2011
|
|
8V11
 
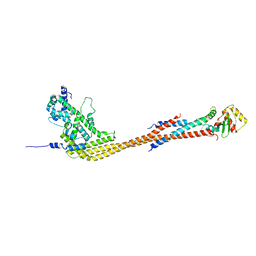 | |
8V10
 
 | |
2I3S
 
 | | Bub3 complex with Bub1 GLEBS motif | | Descriptor: | Cell cycle arrest protein, Checkpoint serine/threonine-protein kinase | | Authors: | Larsen, N.A, Harrison, S.C. | | Deposit date: | 2006-08-20 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of Bub3 interactions in the mitotic spindle checkpoint.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I3T
 
 | | Bub3 complex with Mad3 (BubR1) GLEBS motif | | Descriptor: | Cell cycle arrest protein, Spindle assembly checkpoint component | | Authors: | Larsen, N.A, Harrison, S.C. | | Deposit date: | 2006-08-20 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of Bub3 interactions in the mitotic spindle checkpoint.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8G0P
 
 | |
8G0Q
 
 | |
7UMK
 
 | | Structure of vesicular stomatitis virus (helical reconstruction, 4.1 A resolution) | | Descriptor: | Matrix protein, Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Jenni, S, Horwitz, J.A, Bloyet, L.-M, Whelan, S.P.J, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle.
Nat Commun, 13, 2022
|
|
7UML
 
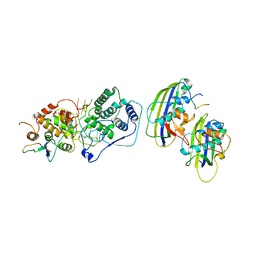 | | Structure of vesicular stomatitis virus (local reconstruction, 3.5 A resolution) | | Descriptor: | Matrix protein, Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Jenni, S, Horwitz, J.A, Bloyet, L.-M, Whelan, S.P.J, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle.
Nat Commun, 13, 2022
|
|
7KQH
 
 | |
7UMT
 
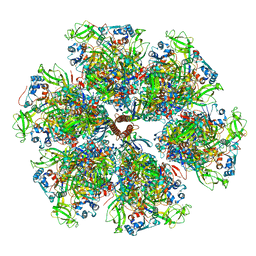 | | Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 - Reversed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Jenni, S, Zongli, L, Wang, Y, Bessey, T, Salgado, E.N, Schmidt, A.G, Greenberg, H.B, Jiang, B, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.
J.Virol., 96, 2022
|
|
7UMS
 
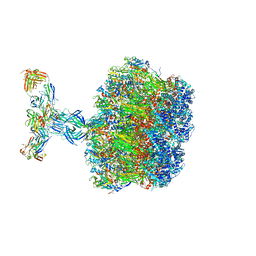 | | Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 in complex with antibody 41 - Upright conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fab 41 heavy chain, ... | | Authors: | Jenni, S, Zongli, L, Wang, Y, Bessey, T, Salgado, E.N, Schmidt, A.G, Greenberg, H.B, Jiang, B, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.
J.Virol., 96, 2022
|
|
2IUH
 
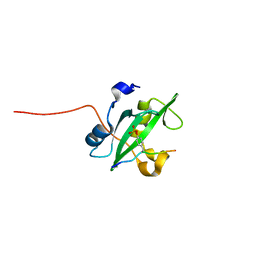 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with c-Kit phosphotyrosyl peptide | | Descriptor: | C-KIT PHOSPHOTYROSYL PEPTIDE, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUG
 
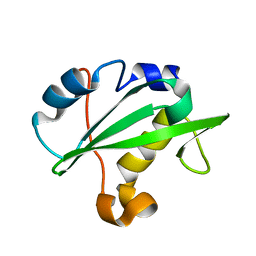 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUI
 
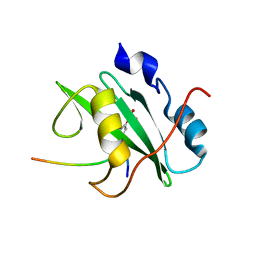 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with PDGFR phosphotyrosyl peptide | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Platelet-derived growth factor receptor beta | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino- Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
7SWO
 
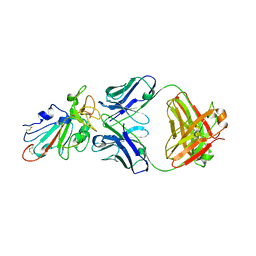 | | C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | C98C7 Fab heavy chain, C98C7 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWP
 
 | | G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32Q4 Fab heavy chain, G32Q4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWN
 
 | | G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32A4 Fab heavy chain, G32A4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
5KTB
 
 | |
6NUW
 
 | | Yeast Ctf19 complex | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2019-02-03 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The structure of the Ctf19c/CCAN from budding yeast.
Elife, 8, 2019
|
|
2BF1
 
 | | Structure of an unliganded and fully-glycosylated SIV gp120 envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, B, Vogan, E.M, Gong, H, Skehel, J.J, Wiley, D.C, Harrison, S.C. | | Deposit date: | 2004-12-02 | | Release date: | 2005-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of an Unliganded Simian Immunodeficiency Virus Gp120 Core
Nature, 433, 2005
|
|
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
