3D31
 
 | | ModBC from Methanosarcina acetivorans | | Descriptor: | Sulfate/molybdate ABC transporter, ATP-binding protein, permease protein, ... | | Authors: | Gerber, S, Comellas-Bigler, M, Locher, K.P. | | Deposit date: | 2008-05-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter.
Science, 321, 2008
|
|
2CTV
 
 | |
5C76
 
 | |
3RCE
 
 | | Bacterial oligosaccharyltransferase PglB | | Descriptor: | MAGNESIUM ION, Oligosaccharide transferase to N-glycosylate proteins, Substrate Mimic Peptide | | Authors: | Lizak, C, Gerber, S, Numao, S, Aebi, M, Locher, K.P. | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray structure of a bacterial oligosaccharyltransferase.
Nature, 474, 2011
|
|
1CON
 
 | | THE REFINED STRUCTURE OF CADMIUM SUBSTITUTED CONCANAVALIN A AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CADMIUM ION, CALCIUM ION, CONCANAVALIN A | | Authors: | Naismith, J.H, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Kalb(Gilboa), A.J, Yariv, J, Wan, T.C.M, Weisgerber, S. | | Deposit date: | 1993-03-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of cadmium-substituted concanavalin A at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
2YPN
 
 | | Hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PROTEIN (HYDROXYMETHYLBILANE SYNTHASE) | | Authors: | Nieh, Y.P, Raftery, J, Weisgerber, S, Habash, J, Schotte, F, Ursby, T, Wulff, M, Haedener, A, Campbell, J.W, Hao, Q, Helliwell, J.R. | | Deposit date: | 1999-01-11 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Accurate and highly complete synchrotron protein crystal Laue diffraction data using the ESRF CCD and the Daresbury Laue software
J.Synchrotron Radiat., 6, 1999
|
|
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q95
 
 | | Crystal structure of the SARS-CoV-2 BA.1 RBD with neutralizing-VHHs Ma16B06 and Ma3F05 | | Descriptor: | Nanobody Ma16B06, Nanobody Ma3F05, Spike protein S1 | | Authors: | Aksu, M, Rymarenko, O, Guttler, T, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q94
 
 | |
8Q93
 
 | | Crystal structure of the SARS-COV-2 RBD with neutralizing-VHHs Re30H02 and Re21D01 | | Descriptor: | Nanobody Re21D01, Nanobody Re30H02, Spike protein S1 | | Authors: | Aksu, M, Guttler, T, Rymarenko, O, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
5C73
 
 | |
5C78
 
 | |
5CNA
 
 | | REFINED STRUCTURE OF CONCANAVALIN A COMPLEXED WITH ALPHA-METHYL-D-MANNOPYRANOSIDE AT 2.0 ANGSTROMS RESOLUTION AND COMPARISON WITH THE SACCHARIDE-FREE STRUCTURE | | Descriptor: | CALCIUM ION, CHLORIDE ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Emmerich, C, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Raftery, J, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1994-02-11 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of concanavalin A complexed with methyl alpha-D-mannopyranoside at 2.0 A resolution and comparison with the saccharide-free structure.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1NLS
 
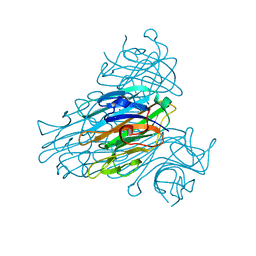 | | CONCANAVALIN A AND ITS BOUND SOLVENT AT 0.94A RESOLUTION | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Deacon, A.M, Gleichmann, T, Helliwell, J.R, Kalb(Gilboa), A.J. | | Deposit date: | 1997-01-28 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Structure of Concanavalin a and its Bound Solvent Determined with Small-Molecule Accuracy at 0.94 A Resolution
J.Chem.Soc.,Faraday Trans., 93, 1997
|
|
1C57
 
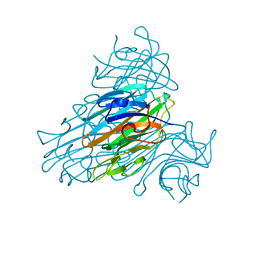 | | DIRECT DETERMINATION OF THE POSITIONS OF DEUTERIUM ATOMS OF BOUND WATER IN CONCANAVALIN A BY NEUTRON LAUE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, Concanavalin-Br, MANGANESE (II) ION | | Authors: | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb, A.J, Helliwell, J.R. | | Deposit date: | 1999-10-26 | | Release date: | 2000-05-08 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Direct determination of the positions of the deuterium atoms of the bound water in -concanavalin A by neutron Laue crystallography.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1CVN
 
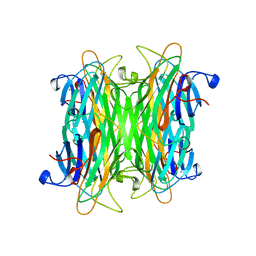 | | CONCANAVALIN A COMPLEXED TO TRIMANNOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Naismith, J.H. | | Deposit date: | 1995-08-09 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of trimannoside recognition by concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1QNY
 
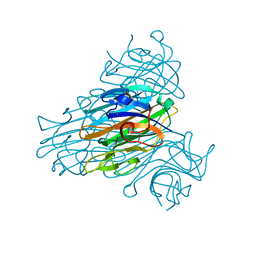 | | X-ray refinement of D2O soaked crystal of concanavalin A | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb(Gilboa), A.J, Helliwell, J.R. | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct Determination of the Positions of Deuterium Atoms of Bound Water in Concanavalin a by Neutron Laue Crystallography
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1SCR
 
 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, CONCANAVALIN A, NICKEL (II) ION | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1SCS
 
 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, COBALT (II) ION, CONCANAVALIN A | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
