4O5R
 
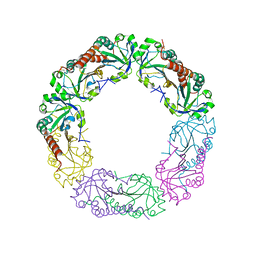 | | Crystal structure of Alkylhydroperoxide Reductase subunit C from E. coli | | Descriptor: | AhpC component, subunit of alkylhydroperoxide reductase | | Authors: | Dip, P.V, Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O5Q
 
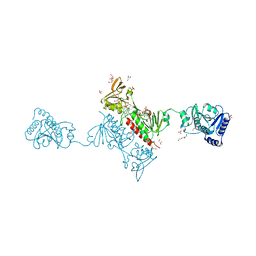 | | Crystal Structure of the Alkylhydroperoxide Reductase AhpF from Escherichia coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkyl hydroperoxide reductase subunit F, CADMIUM ION, ... | | Authors: | Dip, P.V, Kamariah, N, Manimekalai, M.S.S, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O5U
 
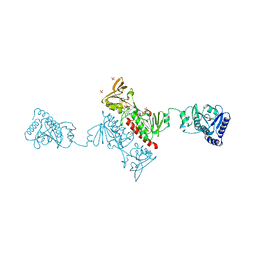 | | Crystal structure of Alkylhydroperoxide Reductase subunit F from E. coli at 2.65 Ang resolution | | Descriptor: | Alkyl hydroperoxide reductase subunit F, CADMIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kamariah, N, Dip, P.V, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2013-12-20 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8VI4
 
 | | TehA from Haemophilus influenzae purified in LMNG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI3
 
 | | TehA from Haemophilus influenzae purified in GDN | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI5
 
 | | TehA from Haemophilus influenzae purified in OG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI2
 
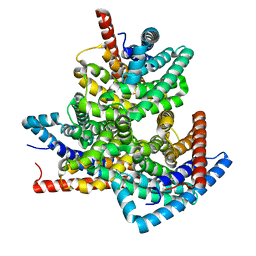 | | TehA from Haemophilus influenzae purified in DDM | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VAF
 
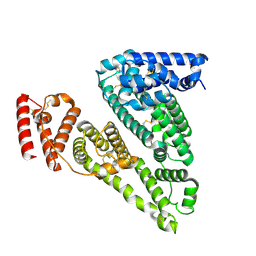 | | Cryogenic electron microscopy structure of apo human serum albumin | | Descriptor: | Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
8VAE
 
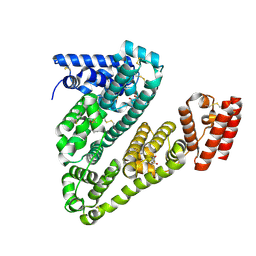 | | Cryogenic electron microscopy structure of human serum albumin in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
8VAC
 
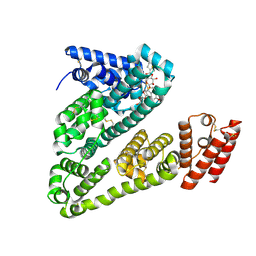 | | Cryogenic electron microscopy structure of human serum albumin in complex with teniposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
4QL9
 
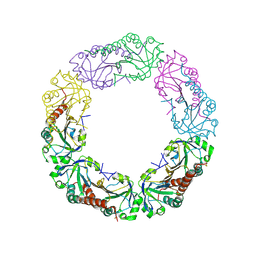 | |
4QL7
 
 | | Crystal structure of C-terminus truncated Alkylhydroperoxide Reductase subunit C (AhpC1-172) from E. coli | | Descriptor: | Alkylhydroperoxide Reductase subunit C | | Authors: | Kamariah, N, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2014-06-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Key roles of the Escherichia coli AhpC C-terminus in assembly and catalysis of alkylhydroperoxide reductase, an enzyme essential for the alleviation of oxidative stress.
Biochim.Biophys.Acta, 1837, 2014
|
|
5B8A
 
 | | Crystal structure of oxidized chimeric EcAhpC1-186-YFSKHN | | Descriptor: | Alkyl hydroperoxide reductase subunit C,Peroxiredoxin-2, GLYCEROL, SULFATE ION | | Authors: | Kamariah, N, Sek, M.F, Eisenhaber, B, Eisenhaber, F, Gruber, G. | | Deposit date: | 2016-06-14 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transition steps in peroxide reduction and a molecular switch for peroxide robustness of prokaryotic peroxiredoxins.
Sci Rep, 6, 2016
|
|
5B8B
 
 | | Crystal structure of reduced chimeric E.coli AhpC1-186-YFSKHN | | Descriptor: | Alkyl hydroperoxide reductase subunit C,Peroxiredoxin-2, SULFATE ION | | Authors: | Kamariah, N, Sek, M.F, Eisenhaber, B, Eisenhaber, F, Gruber, G. | | Deposit date: | 2016-06-14 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Transition steps in peroxide reduction and a molecular switch for peroxide robustness of prokaryotic peroxiredoxins.
Sci Rep, 6, 2016
|
|
2LX4
 
 | |
5Y63
 
 | | Crystal structure of Enterococcus faecalis AhpC | | Descriptor: | Alkyl hydroperoxide reductase, C subunit | | Authors: | Pan, A, Balakrishna, A.M, Grueber, G. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Atomic structure and enzymatic insights into the vancomycin-resistant Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit C
Free Radic. Biol. Med., 115, 2017
|
|
4YKG
 
 | | Crystal Structure of the Alkylhydroperoxide Reductase subunit F (AhpF) with NAD+ from Escherichia coli | | Descriptor: | Alkyl hydroperoxide reductase subunit F, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps
Biochim.Biophys.Acta, 1847, 2015
|
|
4YKF
 
 | | Crystal Structure of the Alkylhydroperoxide Reductase subunit F (AhpF) with NADH from Escherichia coli | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alkyl hydroperoxide reductase subunit F, CADMIUM ION, ... | | Authors: | Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps
Biochim.Biophys.Acta, 1847, 2015
|
|
7PER
 
 | | Model of the inner ring of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup155, Nuclear pore complex protein Nup205, Nuclear pore complex protein Nup93, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
